
Homogentisic acid is a phenolic acid usually found in Arbutus unedo (strawberry-tree) honey. It is also present in the bacterial plant pathogen Xanthomonas campestris pv. phaseoli as well as in the yeast Yarrowia lipolytica where it is associated with the production of brown pigments. It is oxidatively dimerised to form hipposudoric acid, one of the main constituents of the 'blood sweat' of hippopotamuses.
Pyruvate dehydrogenase (NADP+) EC 1.2.1.51 is an enzyme that should not be confused with Pyruvate dehydrogenase (acetyltransferase) EC 1.2.4.1.

Catechol 1,2- dioxygenase is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:
Phytotoxins are substances that are poisonous or toxic to the growth of plants. Phytotoxic substances may result from human activity, as with herbicides, or they may be produced by plants, by microorganisms, or by naturally occurring chemical reactions.

Pseudomonas syringae is a rod-shaped, Gram-negative bacterium with polar flagella. As a plant pathogen, it can infect a wide range of species, and exists as over 50 different pathovars, all of which are available to researchers from international culture collections such as the NCPPB, ICMP, and others.

Pseudomonas savastanoi is a gram-negative plant pathogenic bacterium that infects a variety of plants. It was once considered a pathovar of Pseudomonas syringae, but following DNA-relatedness studies, it was instated as a new species. It is named after Savastano, a worker who proved between 1887 and 1898 that olive knot are caused by bacteria.

Halo blight of bean is a bacterial disease caused by Pseudomonas syringae pv. phaseolicola. Halo blight’s pathogen is a gram-negative, aerobic, polar-flagellated and non-spore forming bacteria. This bacterial disease was first discovered in the early 1920s, and rapidly became the major disease of beans throughout the world. The disease favors the places where temperatures are moderate and plentiful inoculum is available.

Lanosterol synthase is an oxidosqualene cyclase (OSC) enzyme that converts (S)-2,3-oxidosqualene to a protosterol cation and finally to lanosterol. Lanosterol is a key four-ringed intermediate in cholesterol biosynthesis. In humans, lanosterol synthase is encoded by the LSS gene.

N-acetylgalactosamine-6-sulfatase is an enzyme that, in humans, is encoded by the GALNS gene.

Gamma-butyrobetaine dioxygenase is an enzyme that in humans is encoded by the BBOX1 gene. Gamma-butyrobetaine dioxygenase catalyses the formation of L-carnitine from gamma-butyrobetaine, the last step in the L-carnitine biosynthesis pathway. Carnitine is essential for the transport of activated fatty acids across the mitochondrial membrane during mitochondrial beta oxidation. In humans, gamma-butyrobetaine dioxygenase can be found in the kidney (high), liver (moderate), and brain. BBOX1 has recently been identified as a potential cancer gene based on a large-scale microarray data analysis.
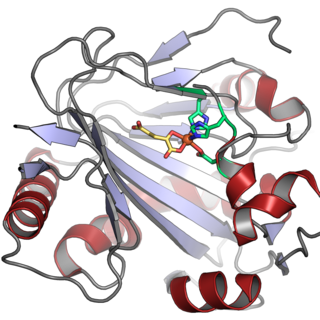
In enzymology, a phytanoyl-CoA dioxygenase (EC 1.14.11.18) is an enzyme that catalyzes the chemical reaction

Procollagen-proline dioxygenase, commonly known as prolyl hydroxylase, is a member of the class of enzymes known as alpha-ketoglutarate-dependent hydroxylases. These enzymes catalyze the incorporation of oxygen into organic substrates through a mechanism that requires alpha-Ketoglutaric acid, Fe2+, and ascorbate. This particular enzyme catalyzes the formation of (2S, 4R)-4-hydroxyproline, a compound that represents the most prevalent post-translational modification in the human proteome.
In enzymology, a taurine dioxygenase (EC 1.14.11.17) is an enzyme that catalyzes the chemical reaction.
In enzymology, a 3,4-dihydroxyphenylacetate 2,3-dioxygenase (EC 1.13.11.15) is an enzyme that catalyzes the chemical reaction

Endothelin receptor type B, (ET-B) is a protein that in humans is encoded by the EDNRB gene.
In enzymology, a D-amino-acid transaminase is an enzyme that catalyzes the chemical reaction:

Glycine cleavage system H protein, mitochondrial is a protein that in humans is encoded by the GCSH gene. Degradation of glycine is brought about by the glycine cleavage system (GCS), which is composed of 4 protein components: P protein, H protein, T protein, and L protein. The H protein shuttles the methylamine group of glycine from the P protein to the T protein. The protein encoded by GCSH gene is the H protein, which transfers the methylamine group of glycine from the P protein to the T protein. Defects in this gene are a cause of nonketotic hyperglycinemia (NKH). Two transcript variants, one protein-coding and the other probably not protein-coding, have been found for this gene. Also, several transcribed and non-transcribed pseudogenes of this gene exist throughout the genome.
Hypoxia-inducible factor-proline dioxygenase (EC 1.14.11.29, HIF hydroxylase) is an enzyme with systematic name hypoxia-inducible factor-L-proline, 2-oxoglutarate:oxygen oxidoreductase (4-hydroxylating). This enzyme catalyses the following chemical reaction
2-oxoglutarate/L-arginine monooxygenase/decarboxylase (succinate-forming) (EC 1.14.11.34, ethylene-forming enzyme, EFE) is an enzyme with systematic name L-arginine,2-oxoglutarate:oxygen oxidoreductase (succinate-forming). This enzyme catalyses the following chemical reaction

Aminopeptidase Y is an enzyme. This enzyme catalyses the following chemical reaction












