Aromatic-ring-hydroxylating dioxygenases (ARHD) incorporate two atoms of dioxygen (O2) into their substrates in the dihydroxylation reaction. The product is (substituted) cis-1,2-dihydroxycyclohexadiene, which is subsequently converted to (substituted) benzene glycol by a cis-diol dehydrogenase.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.

Catechol 2,3-dioxygenase (EC 1.13.11.2, 2,3-pyrocatechase, catechol 2,3-oxygenase, catechol oxygenase, metapyrocatechase, pyrocatechol 2,3-dioxygenase) is an enzyme with systematic name catechol:oxygen 2,3-oxidoreductase (decyclizing). This enzyme catalyses the following chemical reaction

Catechol 1,2- dioxygenase is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:
In enzymology, a 1,6-dihydroxycyclohexa-2,4-diene-1-carboxylate dehydrogenase (EC 1.3.1.25) is an enzyme that catalyzes the chemical reaction
In enzymology, a cis-1,2-dihydrobenzene-1,2-diol dehydrogenase (EC 1.3.1.19) is an enzyme that catalyzes the chemical reaction
In enzymology, a cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (EC 1.3.1.56) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2-chlorobenzoate 1,2-dioxygenase (EC 1.14.12.13) is an enzyme that catalyzes the chemical reaction

In enzymology, a benzene 1,2-dioxygenase is an enzyme that catalyzes the chemical reaction
In enzymology, a toluene dioxygenase (EC 1.14.12.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2,3-dihydroxybenzoate 2,3-dioxygenase (EC 1.13.11.28) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2,3-dihydroxybenzoate 3,4-dioxygenase (EC 1.13.11.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-carboxyethylcatechol 2,3-dioxygenase (EC 1.13.11.16) is an enzyme that catalyzes the chemical reaction
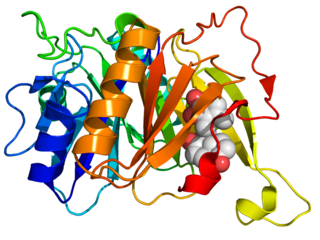
Biphenyl-2,3-diol 1,2-dioxygenase (EC 1.13.11.39) is an enzyme that catalyzes the chemical reaction
Chloridazon-catechol dioxygenase (EC 1.13.11.36) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydroxyquinol 1,2-dioxygenase (EC 1.13.11.37) is an enzyme that catalyzes the chemical reaction
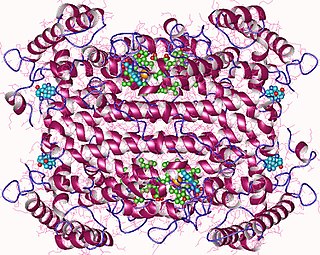
In enzymology, tryptophan 2,3-dioxygenase (EC 1.13.11.11) is a heme enzyme that catalyzes the oxidation of L-tryptophan (L-Trp) to N-formyl-L-kynurenine, as the first and rate-limiting step of the kynurenine pathway.
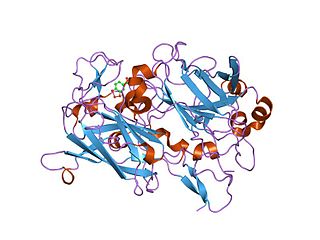
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine triphosphate (ATP) generation to xenobiotic degradation, the use of dioxygen as a biological oxidant is widespread and varied in the exact mechanism of its use. Enzymes employ many different schemes to use dioxygen, and this largely depends on the substrate and reaction at hand.
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Lawrence Que Jr. is a chemist who specializes in bioinorganic chemistry and is a Regents Professor at the University of Minnesota, Twin Cities. He received the 2017 American Chemical Society (ACS) Award in Inorganic Chemistry for his contributions to the field., and the 2008 ACS Alfred Bader Award in Bioinorganic Chemistry.







