
Pseudomonas is a genus of Gram-negative bacteria belonging to the family Pseudomonadaceae in the class Gammaproteobacteria. The 313 members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.

Pseudomonas fluorescens is a common Gram-negative, rod-shaped bacterium. It belongs to the Pseudomonas genus; 16S rRNA analysis as well as phylogenomic analysis has placed P. fluorescens in the P. fluorescens group within the genus, to which it lends its name.

Burkholderia is a genus of Pseudomonadota whose pathogenic members include the Burkholderia cepacia complex, which attacks humans and Burkholderia mallei, responsible for glanders, a disease that occurs mostly in horses and related animals; Burkholderia pseudomallei, causative agent of melioidosis; and Burkholderia cepacia, an important pathogen of pulmonary infections in people with cystic fibrosis (CF). Burkholderia species is also found in marine environments. S.I. Paul et al. (2021) isolated and characterized Burkholderia cepacia from marine sponges of the Saint Martin's Island of the Bay of Bengal, Bangladesh.

The Pseudomonadaceae are a family of bacteria which includes the genera Azomonas, Azorhizophilus, Azotobacter, Mesophilobacter, Pseudomonas, and Rugamonas. The family Azotobacteraceae was recently reclassified into this family.
Pseudomonas chlororaphis is a bacterium used as a soil inoculant in agriculture and horticulture. It can act as a biocontrol agent against certain fungal plant pathogens via production of phenazine-type antibiotics. Based on 16S rRNA analysis, similar species have been placed in its group.

Rhodococcus is a genus of aerobic, nonsporulating, nonmotile Gram-positive bacteria closely related to Mycobacterium and Corynebacterium. While a few species are pathogenic, most are benign, and have been found to thrive in a broad range of environments, including soil, water, and eukaryotic cells. Some species have large genomes, including the 9.7 megabasepair genome of Rhodococcus sp. RHA1.
Pseudomonas veronii is a Gram-negative, rod-shaped, fluorescent, motile bacterium isolated from natural springs in France. It may be used for bioremediation of contaminated soils, as it has been shown to degrade a variety of simple aromatic organic compounds. Based on 16S rRNA analysis, P. veronii has been placed in the P. fluorescens group.

Pseudomonas gessardii is a fluorescent, Gram-negative, rod-shaped bacterium isolated from natural mineral waters in France. Based on 16S rRNA analysis, P. gessardii has been placed in the P. fluorescens group.
Pseudomonas stutzeri is a Gram-negative soil bacterium that is motile, has a single polar flagellum, and is classified as bacillus, or rod-shaped. While this bacterium was first isolated from human spinal fluid, it has since been found in many different environments due to its various characteristics and metabolic capabilities. P. stutzeri is an opportunistic pathogen in clinical settings, although infections are rare. Based on 16S rRNA analysis, this bacterium has been placed in the P. stutzeri group, to which it lends its name.
Paucimonas lemoignei, formerly [Pseudomonas lemoignei], is a Gram-negative soil bacterium. It is aerobic, motile, and rod-shaped.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.
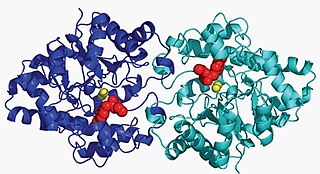
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:

Viable but nonculturable (VBNC) bacteria refers as to bacteria that are in a state of very low metabolic activity and do not divide, but are alive and have the ability to become culturable once resuscitated.
Bacillus halodurans is a rod-shaped, Gram-positive, motile and spore-forming bacterium found in soil. In a genomic comparison with Bacillus subtilis, B. halodurans strain C-125 - originally an unclassified Bacillus strain - was found to contain unique genes and sigma factors that may have aided its adaptation to more alkaline environments.
Alkalihalobacillus alcalophilus is a Gram-positive, rod-shaped species of bacteria. Likely strains of this species have been isolated from highly alkaline waste water. A. alcalophilus is a moderate halotolerant obligate alkaliphile growing at 40 °C and at pH 9–10.5 that has been isolated from soil and animal manures.
Pseudomonas protegens are widespread Gram-negative, plant-protecting bacteria. Some of the strains of this novel bacterial species previously belonged to P. fluorescens. They were reclassified since they seem to cluster separately from other fluorescent Pseudomonas species. P. protegens is phylogenetically related to the Pseudomonas species complexes P. fluorescens, P. chlororaphis, and P. syringae. The bacterial species characteristically produces the antimicrobial compounds pyoluteorin and 2,4-diacetylphloroglucinol (DAPG) which are active against various plant pathogens.
Rhodococcus erythropolis is an aerobic Gram-positive bacterium species in the genus Rhodococcus. The name Rhodococcus erythropolis is derived from its morphogenetic cycle from branching to rod and to coccus morphology, which explains the series of morphological changes this bacterium undergoing during growth and development processes. These bacterium are found in red and orange colonies when observed this explains the species name erythropolis which means "red city" in Greek.
Sphingobium indicum is a hexachlorocyclohexane-degrading bacteria with type strain MTCC 6364T. Its genome has been sequenced.
Intrasporangium is a genus of Gram positive, nonmotile bacteria. The genus name refers to the mycelium of the type strain forming intercalary vesicles that were originally identified as spores. However, no spores have been observed in later studies. The family Intrasporangiaceae is named after the genus, and Intrasporangium is the type genus for the family.

Plastic degradation in marine bacteria describes when certain pelagic bacteria break down polymers and use them as a primary source of carbon for energy. Polymers such as polyethylene (PE), polypropylene (PP), and polyethylene terephthalate (PET) are incredibly useful for their durability and relatively low cost of production, however it is their persistence and difficulty to be properly disposed of that is leading to pollution of the environment and disruption of natural processes. It is estimated that each year there are 9-14 million metric tons of plastic that are entering the ocean due to inefficient solutions for their disposal. The biochemical pathways that allow for certain microbes to break down these polymers into less harmful byproducts has been a topic of study to develop a suitable anti-pollutant.










