Related Research Articles

Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species.

Saccharomyces cerevisiae is a species of yeast. The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. It is the microorganism behind the most common type of fermentation. S. cerevisiae cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.
β-Fructofuranosidase is an enzyme that catalyzes the hydrolysis (breakdown) of the table sugar sucrose into fructose and glucose. Alternative names for β-fructofuranosidase EC 3.2.1.26 include invertase, saccharase, glucosucrase, β-fructosidase, invertin, sucrase, fructosylinvertase, alkaline invertase, acid invertase, and the systematic name: β-fructofuranosidase. The resulting mixture of fructose and glucose is called inverted sugar syrup. Related to invertases are sucrases. Invertases and sucrases hydrolyze sucrose to give the same mixture of glucose and fructose. Invertase is a glycoprotein that hydrolyses (cleaves) the non-reducing terminal β-fructofuranoside residues. Invertases cleave the O-C(fructose) bond, whereas the sucrases cleave the O-C(glucose) bond. Invertase cleaves the α-1,2-glycosidic bond of sucrose.
Saccharomyces boulardii is a tropical yeast first isolated from lychee and mangosteen fruit peel in 1923 by French scientist Henri Boulard. Although early reports claimed distinct taxonomic, metabolic, and genetic properties, S. boulardii is genetically a grouping of S. cerevisiae strains, sharing >99% genomic relatedness, giving the synonym S. cerevisiae var. boulardii.

Glycerol-3-phosphate dehydrogenase (GPDH) is an enzyme that catalyzes the reversible redox conversion of dihydroxyacetone phosphate to sn-glycerol 3-phosphate.
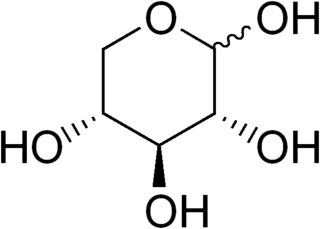
D-Xylose is a five-carbon aldose that can be catabolized or metabolized into useful products by a variety of organisms.
Kexin is a prohormone-processing protease, specifically a yeast serine peptidase, found in the budding yeast. It catalyzes the cleavage of -Lys-Arg- and -Arg-Arg- bonds to process yeast alpha-factor pheromone and killer toxin precursors. The human homolog is PCSK4. It is a family of subtilisin-like peptidases. Even though there are a few prokaryote kexin-like peptidases, all kexins are eukaryotes. The enzyme is encoded by the yeast gene KEX2, and usually referred to in the scientific community as Kex2p. It shares structural similarities with the bacterial protease subtilisin. The first mammalian homologue of this protein to be identified was furin. In the mammal, kexin-like peptidases function in creating and regulating many differing proproteins.
In enzymology, an initiation-specific alpha-1,6-mannosyltransferase is an enzyme that catalyzes the chemical reaction in which an alpha-D-mannosyl residue is transferred from GDP-mannose to a lipid-linked oligosaccharide, being linked by an alpha-1,6-D-mannosyl-D-mannose bond.
Diacetyl reductase ((R)-acetoin forming) (EC 1.1.1.303, (R)-acetoin dehydrogenase) is an enzyme with systematic name (R)-acetoin:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Multisite-specific tRNA:(cytosine-C5)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (cytosine-C5)-methyltransferase. This enzyme catalyses the following chemical reaction
Very-long-chain 3-oxoacyl-CoA synthase (EC 2.3.1.199, very-long-chain 3-ketoacyl-CoA synthase, very-long-chain beta-ketoacyl-CoA synthase, condensing enzyme, CUT1 (gene), CERS6 (gene), FAE1 (gene), KCS (gene), ELO (gene)) is an enzyme with systematic name malonyl-CoA:very-long-chain acyl-CoA malonyltransferase (decarboxylating and thioester-hydrolysing). This enzyme catalyses the following chemical reaction
Ditrans,polycis-polyprenyl diphosphate synthase is an enzyme with systematic name (2E,6E)-farnesyl-diphosphate:isopentenyl-diphosphate cistransferase . This enzyme catalyses the following chemical reaction
Carboxypeptidase D can refer to one of several enzymes. A family of serine carboxypeptidases includes is an enzyme. This enzyme has an optimal pH of 4.5-6.0, is inhibited by diisopropyl fluorophosphate, and catalyses the following chemical reaction
Cerevisin is an enzyme. This enzyme catalyses the following chemical reaction
Ulp1 peptidase is an enzyme. This enzyme catalyses the following chemical reaction
Saccharopepsin is an enzyme. This enzyme catalyses the following chemical reaction
Saccharolysin is an enzyme. This enzyme catalyses the following chemical reaction
Aerobic fermentation or aerobic glycolysis is a metabolic process by which cells metabolize sugars via fermentation in the presence of oxygen and occurs through the repression of normal respiratory metabolism. It is referred to as the Crabtree effect in yeast. and is part of the Warburg effect in tumor cells. While aerobic fermentation does not produce adenosine triphosphate (ATP) in high yield, it allows proliferating cells to convert nutrients such as glucose and glutamine more efficiently into biomass by avoiding unnecessary catabolic oxidation of such nutrients into carbon dioxide, preserving carbon-carbon bonds and promoting anabolism.
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast Saccharomyces cerevisiae e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3.
ERG5 or Sterol 22-desaturase is a cytochrome P450 enzyme in the ergosterol biosynthesis pathway of fungi Saccharomyces cerevisiae, with the CYP Symbol CYP61A1. CYP61A1 is one of only three P450 enzyme found in baker's yeast, the other two are CYP51F1 and CYP56A1. The ortholog in Schizosaccharomyces pombe, was named CYP61A3 for historical reasons, and is only one of two P450 enzyme found with CYP51F1. ERG5 catalyzes the C22-C23 double bond formation on the sterol side chain of ergostatrienol to convert it into ergostatetraenol, then the C24 double bond of ergostatetrenol will be hydrogenation reduced into ergosterol by ERG4.
References
- ↑ MacKay VL, Welch SK, Insley MY, Manney TR, Holly J, Saari GC, Parker ML (January 1988). "The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin". Proceedings of the National Academy of Sciences of the United States of America. 85 (1): 55–9. Bibcode:1988PNAS...85...55M. doi: 10.1073/pnas.85.1.55 . PMC 279480 . PMID 3124102.
- ↑ MacKay VL, Armstrong J, Yip C, Welch S, Walker K, Osborn S, Sheppard P, Forstrom J (1991). "Characterization of the Bar Proteinase, an Extracellular Enzyme from the Yeast Saccharomyces Cerevisiae". Structure and Function of the Aspartic Proteinases. Advances in Experimental Medicine and Biology. Vol. 306. pp. 161–72. doi:10.1007/978-1-4684-6012-4_21. ISBN 978-1-4684-6014-8. PMID 1812704.