Inositol trisphosphate or inositol 1,4,5-trisphosphate abbreviated InsP3 or Ins3P or IP3 is an inositol phosphate signaling molecule. It is made by hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2), a phospholipid that is located in the plasma membrane, by phospholipase C (PLC). Together with diacylglycerol (DAG), IP3 is a second messenger molecule used in signal transduction in biological cells. While DAG stays inside the membrane, IP3 is soluble and diffuses through the cell, where it binds to its receptor, which is a calcium channel located in the endoplasmic reticulum. When IP3 binds its receptor, calcium is released into the cytosol, thereby activating various calcium regulated intracellular signals.
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.

Inositol trisphosphate receptor (InsP3R) is a membrane glycoprotein complex acting as a Ca2+ channel activated by inositol trisphosphate (InsP3). InsP3R is very diverse among organisms, and is necessary for the control of cellular and physiological processes including cell division, cell proliferation, apoptosis, fertilization, development, behavior, learning and memory. Inositol triphosphate receptor represents a dominant second messenger leading to the release of Ca2+ from intracellular store sites. There is strong evidence suggesting that the InsP3R plays an important role in the conversion of external stimuli to intracellular Ca2+ signals characterized by complex patterns relative to both space and time, such as Ca2+ waves and oscillations.
Ryanodine receptors form a class of intracellular calcium channels in various forms of excitable animal tissue like muscles and neurons. There are three major isoforms of the ryanodine receptor, which are found in different tissues and participate in different signaling pathways involving calcium release from intracellular organelles. The RYR2 ryanodine receptor isoform is the major cellular mediator of calcium-induced calcium release (CICR) in animal cells.
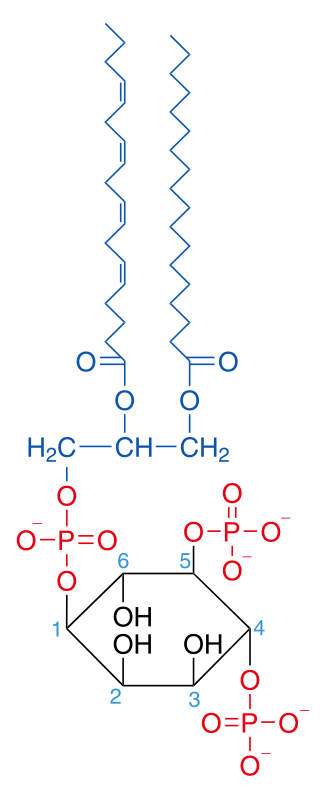
Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins. PIP2 also forms lipid clusters that sort proteins.

Calcium/calmodulin-dependent protein kinase type II subunit alpha (CAMKIIα), a.k.a.Ca2+/calmodulin-dependent protein kinase II alpha, is one subunit of CamKII, a protein kinase (i.e., an enzyme which phosphorylates proteins) that in humans is encoded by the CAMK2A gene.

Inositol 1,4,5-trisphosphate receptor type 1 is a protein that in humans is encoded by the ITPR1 gene.

Calcium/calmodulin-dependent protein kinase type II beta chain is an enzyme that in humans is encoded by the CAMK2B gene.

Synaptotagmin-1 is a protein that in humans is encoded by the SYT1 gene.

Inositol (1,4,5) trisphosphate 3-kinase (EC 2.7.1.127), abbreviated here as ITP3K, is an enzyme that facilitates a phospho-group transfer from adenosine triphosphate to 1D-myo-inositol 1,4,5-trisphosphate. This enzyme belongs to the family of transferases, specifically those transferring phosphorus-containing groups (phosphotransferases) with an alcohol group as acceptor. The systematic name of this enzyme class is ATP:1D-myo-inositol-1,4,5-trisphosphate 3-phosphotransferase. ITP3K catalyzes the transfer of the gamma-phosphate from ATP to the 3-position of inositol 1,4,5-trisphosphate to form inositol 1,3,4,5-tetrakisphosphate. ITP3K is highly specific for the 1,4,5-isomer of IP3, and it exclusively phosphorylates the 3-OH position, producing Ins(1,3,4,5)P4, also known as inositol tetrakisphosphate or IP4.

1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-3 is an enzyme that in humans is encoded by the PLCB3 gene.

1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 is an enzyme that in humans is encoded by the PLCD1 gene. PLCd1 is essential to maintain homeostasis of the skin.
Phospholipase C epsilon 1 (PLCE1) is an enzyme that in humans is encoded by the PLCE1 gene. This gene encodes a phospholipase enzyme (PLCE1) that catalyzes the hydrolysis of phosphatidylinositol-4,5-bisphosphate to generate two second messengers: inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG). Mutations in this gene cause early-onset nephrotic syndrome and have been associated with respiratory chain deficiency with diffuse mesangial sclerosis.

Inositol-trisphosphate 3-kinase A is an enzyme that in humans is encoded by the ITPKA gene.

Calcium/calmodulin-dependent protein kinase kinase 1 is an enzyme that in humans is encoded by the CAMKK1 gene.

Ryanodine receptor 1 (RYR-1) also known as skeletal muscle calcium release channel or skeletal muscle-type ryanodine receptor is one of a class of ryanodine receptors and a protein found primarily in skeletal muscle. In humans, it is encoded by the RYR1 gene.

Inositol 1,4,5-trisphosphate receptor, type 2, also known as ITPR2, is a protein which in humans is encoded by the ITPR2 gene. The protein encoded by this gene is both a receptor for inositol triphosphate and a calcium channel.

Inositol 1,4,5-trisphosphate receptor, type 3, also known as ITPR3, is a protein which in humans is encoded by the ITPR3 gene. The protein encoded by this gene is both a receptor for inositol triphosphate and a calcium channel.
The ryanodine-inositol 1,4,5-triphosphate receptor Ca2+ channel (RIR-CaC) family includes Ryanodine receptors and Inositol trisphosphate receptors. Members of this family are large proteins, some exceeding 5000 amino acyl residues in length. This family belongs to the Voltage-gated ion channel (VIC) superfamily. Ry receptors occur primarily in muscle cell sarcoplasmic reticular (SR) membranes, and IP3 receptors occur primarily in brain cell endoplasmic reticular (ER) membranes where they effect release of Ca2+ into the cytoplasm upon activation (opening) of the channel. They are redox sensors, possibly providing a partial explanation for how they control cytoplasmic Ca2+. Ry receptors have been identified in heart mitochondria where they provide the main pathway for Ca2+ entry. Sun et al. (2011) have demonstrated oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor-Ca2+ release channel (RyR1;TC# 1.A.3.1.2) by NADPH oxidase 4.
Calcium binding protein 2, also known as CaBP2, is a protein that in humans is encoded by the CABP2 gene.



















