
Chloranthaceae is a family of flowering plants (angiosperms), the only family in the order Chloranthales. It is not closely related to any other family of flowering plants, and is among the early-diverging lineages in the angiosperms. They are woody or weakly woody plants occurring in Southeast Asia, the Pacific, Madagascar, Central and South America, and the West Indies. The family consists of four extant genera, totalling about 77 known species according to Christenhusz and Byng in 2016. Some species are used in traditional medicine. The type genus is Chloranthus. The fossil record of the family, mostly represented by pollen such as Clavatipollenites, extends back to the dawn of the history of flowering plants in the Early Cretaceous, and has been found on all continents.
The historical application of biotechnology throughout time is provided below in chronological order.

An artificial enzyme is a synthetic organic molecule or ion that recreates one or more functions of an enzyme. It seeks to deliver catalysis at rates and selectivity observed in naturally occurring enzymes.

Acorus gramineus, commonly known as Japanese sweet flag, Japanese rush, grassy-leaved sweet flag, and grass-leaf sweet flag, is a botanical species belonging to the genus Acorus, native to Japan, Korea, and eastern Asia. The plant usually grows in wetlands and shallow water.

Solute carrier family 52, member 3, formerly known as chromosome 20 open reading frame 54 and riboflavin transporter 2, is a protein that in humans is encoded by the SLC52A3 gene.

François Balloux is the director of the UCL Genetics Institute, and a professor of computational biology at University College London.
The influenza A virus subtype H10N8 is an avian influenza virus. It is one of three H10 subtype avian influenza viruses isolated from domestic ducks in China, designated as SH602/H10N8, FJ1761/H10N3 and SX3180/H10N7.

High-entropy-alloy nanoparticles (HEA-NPs) are nanoparticles having five or more elements alloyed in a single-phase solid solution structure. HEA-NPs possess a wide range of compositional library, distinct alloy mixing structure, and nanoscale size effect, giving them huge potential in catalysis, energy, environmental, and biomedical applications.
Bat coronavirus RaTG13 is a SARS-like betacoronavirus identified in the droppings of the horseshoe bat Rhinolophus affinis. It was discovered in 2013 in bat droppings from a mining cave near the town of Tongguan in Mojiang county in Yunnan, China. In February 2020, it was identified as the closest known relative of SARS-CoV-2, the virus that causes COVID-19, sharing 96.1% nucleotide identity. However, in 2022, scientists found three closer matches in bats found 530 km south, in Feuang, Laos, designated as BANAL-52, BANAL-103 and BANAL-236.
RmYN02 is a bat-derived strain of Severe acute respiratory syndrome–related coronavirus. It was discovered in bat droppings collected between May and October 2019 from sites in Mengla County, Yunnan Province, China. It is the second-closest known relative of SARS-CoV-2, the virus strain that causes COVID-19, sharing 93.3% nucleotide identity at the scale of the complete virus genome. RmYN02 contains an insertion at the S1/S2 cleavage site in the spike protein, similar to SARS-CoV-2, suggesting that such insertion events can occur naturally.
RacCS203 is a bat-derived strain of severe acute respiratory syndrome–related coronavirus collected in acuminate horseshoe bats from sites in Thailand and sequenced by Lin-Fa Wang's team. It has 91.5% sequence similarity to SARS-CoV-2 and is most related to the RmYN02 strain. Its spike protein is closely related to RmYN02's spike, both highly divergent from SARS-CoV-2's spike.
Rc-o319 is a bat-derived strain of severe acute respiratory syndrome–related coronavirus collected in Little Japanese horseshoe bats from sites in Iwate, Japan. Its has 81% similarity to SARS-CoV-2 and is the earliest strain branch of the SARS-CoV-2 related coronavirus.

ORF9b is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is an overlapping gene whose open reading frame is entirely contained within the N gene, which encodes coronavirus nucleocapsid protein. The encoded protein is 97 amino acid residues long in SARS-CoV and 98 in SARS-CoV-2, in both cases forming a protein dimer.
This paleobotany list records new fossil plant taxa that were to be described during the year 2022, as well as notes other significant paleobotany discoveries and events which occurred during 2022.
The human identical sequence (HIS) is a sequence of RNA elements, 24-27 nucleotides in length, that coronavirus genomes share with the human genome. In pathogenic progression, HIS acts as a NamiRNA (nuclear activating miRNA) through the NamiRNA-enhancer network to activate neighboring host genes. The first HIS elements was identified in the SARS-CoV-2 genome, which has five HIS elements; other human coronaviruses have one to five. It has been suggested that these sequences can be more generally termed "host identical sequences" since similar correlations have been found between the genome of SARS-CoV-2 and multiple potential hosts (bats, pangolins, ferrets, and cats).
In photonics, a meta-waveguide is a physical structures that guides electromagnetic waves with engineered functional subwavelength structures. Meta-waveguides are the result of combining the fields of metamaterials and metasurfaces into integrated optics. The design of the subwavelength architecture allows exotic waveguiding phenomena to be explored.

In archaeogenetics, the term Ancient Northern East Asian (ANEA), also known as Northern East Asian (NEA), is used to summarize the related ancestral components that represent the Ancient Northern East Asian peoples, extending from the Baikal region to the Yellow River and the Qinling-Huaihe Line in present-day central China. They are inferred to have diverged from Ancient Southern East Asians (ASEA) around 20,000 to 26,000 BCe.
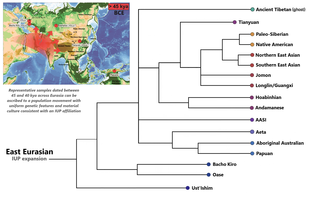
In archaeogenetics, the term Ancient Southern East Asian (ASEA), also known as Southern East Asian (SEA), is used to summarize the related ancestral components that represent the Ancient Southern East Asian peoples, extending from the Fujian region to the coastal Southern China and Taiwan Strait. They are inferred to have diverged from Ancient Northern East Asians (ANEA) around c. 20,000 to 26,000 BCe.
1. Kong, H.-Z., Chen, Z.-D. & Lu, A.-M. (2002) Phylogeny of Chloranthus (Chloranthaceae) based on nuclear ribosomal ITS and plastid TRNL-F sequence data. American Journal of Botany 89(6): 940–946.
2.Li, Q, Y. Wang, S. Wen, Y. Wu, L. Xu, and Z. Sun. 2019. A New Dimeric Sesquiterpenoid from Chloranthus japonicus Sieb. A C G publication 13:483-490









