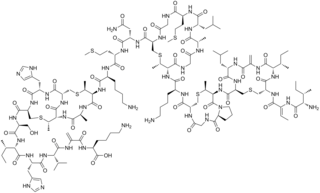
Nisin is a polycyclic antibacterial peptide produced by the bacterium Lactococcus lactis that is used as a food preservative. It has 34 amino acid residues, including the uncommon amino acids lanthionine (Lan), methyllanthionine (MeLan), didehydroalanine (Dha), and didehydroaminobutyric acid (Dhb). These unusual amino acids are introduced by posttranslational modification of the precursor peptide. In these reactions a ribosomally synthesized 57-mer is converted to the final peptide. The unsaturated amino acids originate from serine and threonine, and the enzyme-catalysed addition of cysteine residues to the didehydro amino acids result in the multiple (5) thioether bridges.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.

Clavulanic acid is a β-lactam drug that functions as a mechanism-based β-lactamase inhibitor. While not effective by itself as an antibiotic, when combined with penicillin-group antibiotics, it can overcome antibiotic resistance in bacteria that secrete β-lactamase, which otherwise inactivates most penicillins.
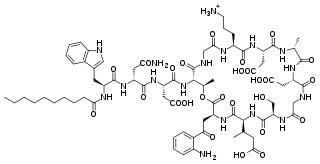
Daptomycin, sold under the brand name Cubicin among others, is a lipopeptide antibiotic used in the treatment of systemic and life-threatening infections caused by Gram-positive organisms.

Geldanamycin is a 1,4-benzoquinone ansamycin antitumor antibiotic that inhibits the function of Hsp90 by binding to the unusual ADP/ATP-binding pocket of the protein. HSP90 client proteins play important roles in the regulation of the cell cycle, cell growth, cell survival, apoptosis, angiogenesis and oncogenesis.

Renin inhibitors are pharmaceutical drugs inhibiting the activity of renin that is responsible for hydrolyzing angiotensinogen to angiotensin I, which in turn reduces the formation of angiotensin II that facilitates blood pressure.

Thiostrepton is a natural cyclic oligopeptide antibiotic of the thiopeptide class, derived from several strains of streptomycetes, such as Streptomyces azureus and Streptomyces laurentii. Thiostrepton is a natural product of the ribosomally synthesized and post-translationally modified peptide (RiPP) class.
Zwittermicin A is an antibiotic that has been identified from the bacterium Bacillus cereus UW85. It is a molecule of interest to agricultural industry because it has the potential to suppress plant disease due to its broad spectrum activity against certain gram positive and gram negative prokaryotic micro-organisms. The molecule is also of interest from a metabolic perspective because it represents a new structural class of antibiotic and suggests a crossover between polyketide and non-ribosomal peptide biosynthetic pathways. Zwittermicin A is linear aminopolyol.
Streptogramin A is a group of antibiotics within the larger family of antibiotics known as streptogramins. They are synthesized by the bacteria Streptomyces virginiae. The streptogramin family of antibiotics consists of two distinct groups: group A antibiotics contain a 23-membered unsaturated ring with lactone and peptide bonds while group B antibiotics are depsipeptides. While structurally different, these two groups of antibiotics act synergistically, providing greater antibiotic activity than the combined activity of the separate components. These antibiotics have until recently been commercially manufactured as feed additives in agriculture, although today there is increased interest in their ability to combat antibiotic-resistant bacteria, particularly vancomycin-resistant bacteria.
Streptogramin B is a subgroup of the streptogramin antibiotics family. These natural products are cyclic hexa- or hepta depsipeptides produced by various members of the genus of bacteria Streptomyces. Many of the members of the streptogramins reported in the literature have the same structure and different names; for example, pristinamycin IA = vernamycin Bα = mikamycin B = osteogrycin B.
Thiazolines are a group of isomeric 5-membered heterocyclic compounds containing both sulfur and nitrogen in the ring. Although unsubstituted thiazolines are rarely encountered themselves, their derivatives are more common and some are bioactive. For example, in a common post-translational modification, cysteine residues are converted into thiazolines.
Plantazolicin (PZN) is a natural antibiotic produced by the gram-positive soil bacterium Bacillus velezensis FZB42. PZN has specifically been identified as a selective bactericidal agent active against Bacillus anthracis, the causative agent of anthrax. This natural product is a ribosomally synthesized and post-translationally modified peptide (RiPP); it can be classified further as a thiazole/oxazole-modified microcin (TOMM) or a linear azole-containing peptide (LAP).

Bottromycin is a macrocyclic peptide with antibiotic activity. It was first discovered in 1957 as a natural product isolated from Streptomyces bottropensis. It has been shown to inhibit methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE) among other Gram-positive bacteria and mycoplasma. Bottromycin is structurally distinct from both vancomycin, a glycopeptide antibiotic, and methicillin, a beta-lactam antibiotic.
Ribosomally synthesized and post-translationally modified peptides (RiPPs), also known as ribosomal natural products, are a diverse class of natural products of ribosomal origin. Consisting of more than 20 sub-classes, RiPPs are produced by a variety of organisms, including prokaryotes, eukaryotes, and archaea, and they possess a wide range of biological functions.
Teixobactin is a peptide-like secondary metabolite of some species of bacteria, that kills some gram-positive bacteria. It appears to belong to a new class of antibiotics, and harms bacteria by binding to lipid II and lipid III, important precursor molecules for forming the cell wall.

Nosiheptide is a thiopeptide antibiotic produced by the bacterium Streptomyces actuosus.

Thiopeptides are a class of peptide antibiotics produced by bacteria. They have antibiotic activity against Gram-positive bacteria, but little or no activity against Gram-negative bacteria. Many of the members of this class show activity against methicillin-resistant Staphylococcus aureus (MRSA) and are therefore subjects of research interest.
Butyrolactol A is an organic chemical compound of interest for its potential use as an antifungal antibiotic.
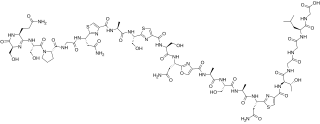
Klebsazolicin (KLB) is a peptide antibiotic encoded in the genome of a gram-negative bacterium Klebsiella pneumoniae subsp. ozonae and targeting prokaryotic ribosome. Klebsazolicin is a ribosomally synthesized and post-translationally modified peptide (RiPP) and a linear azol(in)e-containing peptide (LAP).

Cinnamycin is a tetracyclic antibacterial peptide produced by Streptomyces cinnamoneus containing 19 amino acid residues including the unusual amino acids threo-3-methyl-lanthionine, meso-lanthionine, lysinoalanine, and 3-hydroxyaspartic acid.












