Substrate-level phosphorylation is a metabolism reaction that results in the production of ATP or GTP supported by the energy released from another high-energy bond that leads to phosphorylation of ADP or GDP to ATP or GTP (note that the reaction catalyzed by creatine kinase is not considered as "substrate-level phosphorylation"). This process uses some of the released chemical energy, the Gibbs free energy, to transfer a phosphoryl (PO3) group to ADP or GDP. Occurs in glycolysis and in the citric acid cycle.

10-Formyltetrahydrofolate (10-CHO-THF) is a form of tetrahydrofolate that acts as a donor of formyl groups in anabolism. In these reactions 10-CHO-THF is used as a substrate in formyltransferase reactions.
In enzymology, a formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-methyl-2-oxobutanoate hydroxymethyltransferase (EC 2.1.2.11) is an enzyme that catalyzes the chemical reaction
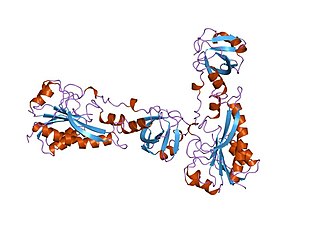
In enzymology, a methionyl-tRNA formyltransferase (EC 2.1.2.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a phosphoribosylaminoimidazolecarboxamide formyltransferase, also known by the shorter name AICAR transformylase, is an enzyme that catalyzes the chemical reaction
In enzymology, a 2-hydroxymuconate-semialdehyde hydrolase (EC 3.7.1.9) is an enzyme that catalyzes the chemical reaction

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction
In enzymology, an arylformamidase (EC 3.5.1.9) is an enzyme that catalyzes the chemical reaction
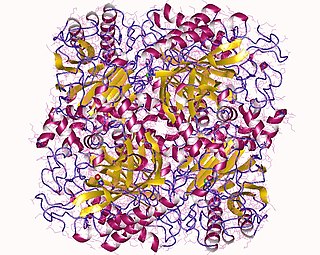
In enzymology, a formamidase (EC 3.5.1.49) is an enzyme that catalyzes the chemical reaction
In enzymology, a formylaspartate deformylase (EC 3.5.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a formylmethionine deformylase (EC 3.5.1.31) is an enzyme that catalyzes the chemical reaction
In enzymology, a GTP cyclohydrolase II (EC 3.5.4.25) is an enzyme that catalyzes the chemical reaction

In enzymology, a methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a N-formylglutamate deformylase (EC 3.5.1.68) is an enzyme that catalyzes the chemical reaction
In enzymology, a N-formylmethionylaminoacyl-tRNA deformylase (EC 3.5.1.27) is an enzyme that catalyzes the chemical reaction
In enzymology, a N,N-dimethylformamidase (EC 3.5.1.56) is an enzyme that catalyzes the chemical reaction
In enzymology, a N-substituted formamide deformylase (EC 3.5.1.91) is an enzyme that catalyzes the chemical reaction

10-formyltetrahydrofolate dehydrogenase is an enzyme that in humans is encoded by the ALDH1L1 gene.

The pfl RNA motif refers to a conserved RNA structure present in some bacteria and originally discovered using bioinformatics. pfl RNAs are consistently present in genomic locations that likely correspond to the 5' untranslated regions of protein-coding genes. This arrangement in bacteria is commonly associated with cis-regulatory elements. Moreover, they are in presumed 5' UTRs of multiple non-homologous genes, suggesting that they function only in these locations. Additional evidence of cis-regulatory function came from the observation that predicted rho-independent transcription terminators overlap pfl RNAs. This overlap suggests that the alternate secondary structures of pfl RNA and the transcription terminator stem-loops compete with each other, and this is a common mechanism for cis gene control in bacteria.







