
Crohn's disease is a type of inflammatory bowel disease (IBD) that may affect any segment of the gastrointestinal tract. Symptoms often include abdominal pain, diarrhea, fever, abdominal distension, and weight loss. Complications outside of the gastrointestinal tract may include anemia, skin rashes, arthritis, inflammation of the eye, and fatigue. The skin rashes may be due to infections as well as pyoderma gangrenosum or erythema nodosum. Bowel obstruction may occur as a complication of chronic inflammation, and those with the disease are at greater risk of colon cancer and small bowel cancer.
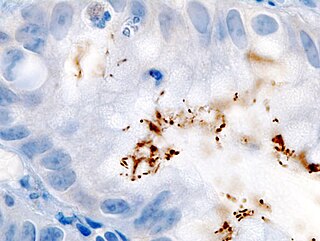
Helicobacter pylori, previously known as Campylobacter pylori, is a gram-negative, microaerophilic, spiral (helical) bacterium usually found in the stomach. Its helical shape is thought to have evolved in order to penetrate the mucoid lining of the stomach and thereby establish infection. The bacterium was first identified in 1982 by the Australian doctors Barry Marshall and Robin Warren. H. pylori has been associated with cancer of the mucosa-associated lymphoid tissue in the stomach, esophagus, colon, rectum, or tissues around the eye, and of lymphoid tissue in the stomach.

Helicobacter is a genus of Gram-negative bacteria possessing a characteristic helical shape. They were initially considered to be members of the genus Campylobacter, but in 1989, Goodwin et al. published sufficient reasons to justify the new genus name Helicobacter. The genus Helicobacter contains about 35 species.

Achlorhydria and hypochlorhydria refer to states where the production of hydrochloric acid in gastric secretions of the stomach and other digestive organs is absent or low, respectively. It is associated with various other medical problems.

NlaIII is a type II restriction enzyme isolated from Neisseria lactamica. As part of the restriction modification system, NlaIII is able to prevent foreign DNA from integrating into the host genome by cutting double stranded DNA into fragments at specific sequences. This results in further degradation of the fragmented foreign DNA and prevents it from infecting the host genome.
Chlamydia muridarum is an intracellular bacterial species that at one time belonged to Chlamydia trachomatis. However, C. trachomatis naturally only infects humans and C. muridarum naturally infects only members of the family Muridae.

Toll-like receptor 5, also known as TLR5, is a protein which in humans is encoded by the TLR5 gene. It is a member of the toll-like receptor (TLR) family. TLR5 is known to recognize bacterial flagellin from invading mobile bacteria. It has been shown to be involved in the onset of many diseases, which includes Inflammatory bowel disease. Recent studies have also shown that malfunctioning of TLR5 is likely related to rheumatoid arthritis, osteoclastogenesis, and bone loss. Abnormal TLR5 functioning is related to the onset of gastric, cervical, endometrial and ovarian cancers.

N-formyl peptide receptor 2 (FPR2) is a G-protein coupled receptor (GPCR) located on the surface of many cell types of various animal species. The human receptor protein is encoded by the FPR2 gene and is activated to regulate cell function by binding any one of a wide variety of ligands including not only certain N-Formylmethionine-containing oligopeptides such as N-Formylmethionine-leucyl-phenylalanine (FMLP) but also the polyunsaturated fatty acid metabolite of arachidonic acid, lipoxin A4 (LXA4). Because of its interaction with lipoxin A4, FPR2 is also commonly named the ALX/FPR2 or just ALX receptor.

Peptidoglycan recognition protein 2(PGLYRP2) is an enzyme, N-acetylmuramoyl-L-alanine amidase (NAMLAA), that hydrolyzes bacterial cell wall peptidoglycan and is encoded by the PGLYRP2 gene.
Mouse models of colorectal cancer and intestinal cancer are experimental systems in which mice are genetically manipulated, fed a modified diet, or challenged with chemicals to develop malignancies in the gastrointestinal tract. These models enable researchers to study the onset, progression of the disease, and understand in depth the molecular events that contribute to the development and spread of colorectal cancer. They also provide a valuable biological system, to simulate human physiological conditions, suitable for testing therapeutics.
Campylobacter concisus is a Gram-negative, highly fastidious, mesophilic bacterium that grows under both anaerobic and microaerobic conditions with the presence of hydrogen significantly aiding growth. Motile, with either unipolar or bipolar flagella, the organisms have a characteristic spiral/corkscrew appearance and are oxidase-positive. Although the human oral cavity is the natural colonization site of the bacterium, C. concisus may also colonize the intestinal tract of some individuals. In particular, several studies have reported higher intestinal prevalence of C. concisus in patients with IBD compared to healthy controls, which has led to current speculation of the bacterium's implication in the induction of Crohn's disease.
Helicobacter hepaticus is a bacterium in the Helicobacteraceae family, Campylobacterales order.

Helicobacter bilis is a bacterium in the Helicobacteraceae family, Campylobacterales order. It is a fusiform bacterium with three to 14 multiple bipolar sheathed flagella and periplasmic fibers wrapped around the cell. It is resistant to cephalothin and nalidixic acid, but sensitive to metronidazole. Like Helicobacter hepaticus, it colonizes the bile, liver, and intestine of mice, and is associated with multifocal chronic hepatitis and hepatocellular tumors.
Helicobacter rodentium is a bacterium in the Helicobacteraceae family, Campylobacterales order. It is a spiral-shaped bacterium with a bipolar, single, nonsheathed flagellum. It is resistant to cephalothin and nalidixic acid. Its type strain is MIT 95-1707. Its name refers to the species first being isolated from mice.
Helicobacter muridarum is a bacterium in the Helicobacteraceae family, Campylobacterales order. It is microaerophilic and helical and was first isolated from the intestinal mucosa of rodents, hence its name. It is characterised by the presence of 9 to 11 periplasmic fibers which appear as concentric helical ridges on the surface of each cell. The cells are motile and have bipolar tufts of 10 to 14 sheathed flagella. These bacteria are nutritionally fastidious and physiologically similar to other Helicobacter species and Wolinella succinogenes, but can be differentiated from these organisms by their unique cellular ultrastructure. ST1T is its type strain.
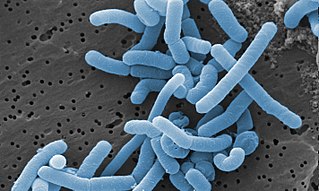
Lacticaseibacillus paracasei is a gram-positive, homofermentative species of lactic acid bacteria that are commonly used in dairy product fermentation and as probiotic cultures. Lc. paracasei is a bacterium that operates by commensalism. It is commonly found in many human habitats such as human intestinal tracts and mouths as well as sewages, silages, and previously mentioned dairy products. The name includes morphology, a rod-shaped bacterium with a width of 2.0 to 4.0μm and length of 0.8 to 1.0μm.

Akkermansia muciniphila is a human intestinal symbiont, isolated from human feces. It is a mucin-degrading bacterium belonging to the genus, Akkermansia, discovered in 2004 by Muriel Derrien and Willem de Vos at Wageningen University of the Netherlands. It belongs to the phylum Verrucomicrobiota and its type strain is MucT. It is under preliminary research for its potential association with metabolic disorders.
The altered Schaedler flora (ASF) is a community of eight bacterial species: two lactobacilli, one Bacteroides, one spiral bacterium of the Flexistipes genus, and four extremely oxygen sensitive (EOS) fusiform-shaped species. The bacteria are selected for their dominance and persistence in the normal microflora of mice, and for their ability to be isolated and grown in laboratory settings. Germ-free animals, mainly mice, are colonized with ASF for the purpose of studying the gastrointestinal (GI) tract. Intestinal mutualistic bacteria play an important role in affecting gene expression of the GI tract, immune responses, nutrient absorption, and pathogen resistance. The standardized microbial cocktail enabled the controlled study of microbe and host interactions, role of microbes, pathogen effects, and intestinal immunity and disease association, such as cancer, inflammatory bowel disease, diabetes, and other inflammatory or autoimmune diseases. Also, compared to germfree animals, ASF mice have fully developed immune system, resistance to opportunistic pathogens, and normal GI function and health, and are a great representation of normal mice.
Akkermansia glycanphila is a species of intestinal mucin-degrading bacterium. It was first isolated from reticulated python feces in 2016.
Helicobacter cetorum is a Gram-negative, microaerophilic, spiral (helical) bacterium that is usually found in the stomachs of whales and dolphins. Based on 16S rRNA sequencing, its genome is very similar to that of Helicobacter pylori in that it can cause gastric disease in these animals. Originally isolated among Atlantic white-sided dolphins and Beluga whales in 2000, H. cetorum has been associated with hemorrhages throughout its entire gastrointestinal tract, but its role has not yet been discovered. Prior to the discovery of H. cetorum, there have not been any other Helicobacter species reported in dolphins.








