The quinones are a class of organic compounds that are formally "derived from aromatic compounds [such as benzene or naphthalene] by conversion of an even number of –CH= groups into –C(=O)– groups with any necessary rearrangement of double bonds, resulting in "a fully conjugated cyclic dione structure". The archetypical member of the class is 1,4-benzoquinone or cyclohexadienedione, often called simply "quinone". Other important examples are 1,2-benzoquinone (ortho-quinone), 1,4-naphthoquinone and 9,10-anthraquinone.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.
Aromatic-ring-hydroxylating dioxygenases (ARHD) incorporate two atoms of dioxygen (O2) into their substrates in the dihydroxylation reaction. The product is (substituted) cis-1,2-dihydroxycyclohexadiene, which is subsequently converted to (substituted) benzene glycol by a cis-diol dehydrogenase.

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.

4-Hydroxyphenylpyruvate dioxygenase (HPPD), also known as α-ketoisocaproate dioxygenase, is an Fe(II)-containing non-heme oxygenase that catalyzes the second reaction in the catabolism of tyrosine - the conversion of 4-hydroxyphenylpyruvate into homogentisate. HPPD also catalyzes the conversion of phenylpyruvate to 2-hydroxyphenylacetate and the conversion of α-ketoisocaproate to β-hydroxy β-methylbutyrate. HPPD is an enzyme that is found in nearly all aerobic forms of life.

Catechol 1,2- dioxygenase is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:
In enzymology, a cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (EC 1.3.1.56) is an enzyme that catalyzes the chemical reaction

In enzymology, a benzene 1,2-dioxygenase is an enzyme that catalyzes the chemical reaction
In enzymology, a biphenyl 2,3-dioxygenase (EC 1.14.12.18) is an enzyme that catalyzes the chemical reaction
In enzymology, a naphthalene 1,2-dioxygenase (EC 1.14.12.12) is an enzyme that catalyzes the chemical reaction
In enzymology, a terephthalate 1,2-dioxygenase (EC 1.14.12.15) is an enzyme that catalyzes the chemical reaction
In enzymology, a toluene dioxygenase (EC 1.14.12.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydroxyquinol 1,2-dioxygenase (EC 1.13.11.37) is an enzyme that catalyzes the chemical reaction
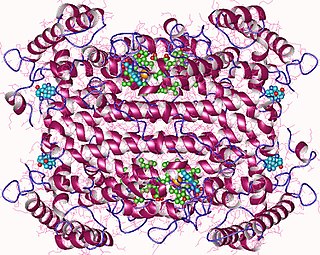
In enzymology, tryptophan 2,3-dioxygenase (EC 1.13.11.11) is a heme enzyme that catalyzes the oxidation of L-tryptophan (L-Trp) to N-formyl-L-kynurenine, as the first and rate-limiting step of the kynurenine pathway.
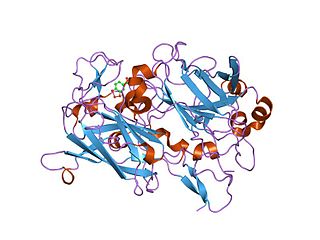
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine triphosphate (ATP) generation to xenobiotic degradation, the use of dioxygen as a biological oxidant is widespread and varied in the exact mechanism of its use. Enzymes employ many different schemes to use dioxygen, and this largely depends on the substrate and reaction at hand.
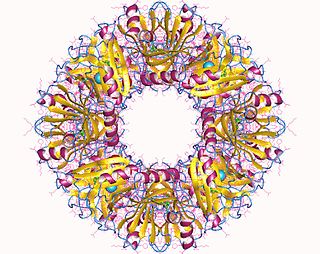
1,2-dihydroxynaphthalene dioxygenase (EC 1.13.11.56, 1,2-DHN dioxygenase, DHNDO, 1,2-dihydroxynaphthalene oxygenase, 1,2-dihydroxynaphthalene:oxygen oxidoreductase) is an enzyme with systematic name naphthalene-1,2-diol:oxygen oxidoreductase. This enzyme catalyses the following chemical reaction
(2,2,3-Trimethyl-5-oxocyclopent-3-enyl)acetyl-CoA 1,5-monooxygenase (EC 1.14.13.160, 2-oxo-Delta3-4,5,5-trimethylcyclopentenylacetyl-CoA monooxygenase, 2-oxo-Delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase, OTEMO) is an enzyme with systematic name ((1R)-2,2,3-trimethyl-5-oxocyclopent-3-enyl)acetyl-CoA,NADPH:oxygen oxidoreductase (1,5-lactonizing). This enzyme catalyses the following chemical reaction
The alpha-D-phosphohexomutases are a large superfamily of enzymes, with members in all three domains of life. Enzymes from this superfamily are ubiquitous in organisms from E. Coli to humans, and catalyze a phosphoryl transfer reaction on a phosphosugar substrate. Four well studied subgroups in the superfamily are:
- Phosphoglucomutase (PGM)
- Phosphoglucomutase/Phosphomannomutase (PGM/PMM)
- Phosphoglucosamine mutase (PNGM)
- Phosphoaceytlglucosamine mutase (PAGM)
Lawrence Que Jr. is a chemist who specializes in bioinorganic chemistry and is a Regents Professor at the University of Minnesota, Twin Cities. He received the 2017 American Chemical Society (ACS) Award in Inorganic Chemistry for his contributions to the field., and the 2008 ACS Alfred Bader Award in Bioinorganic Chemistry.








