Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

Yeast artificial chromosomes (YACs) are genetically engineered chromosomes derived from the DNA of the yeast, Saccharomyces cerevisiae, which is then ligated into a bacterial plasmid. By inserting large fragments of DNA, from 100–1000 kb, the inserted sequences can be cloned and physically mapped using a process called chromosome walking. This is the process that was initially used for the Human Genome Project, however due to stability issues, YACs were abandoned for the use of bacterial artificial chromosome

Synthetic biology (SynBio) is a multidisciplinary field of science that focuses on living systems and organisms, and it applies engineering principles to develop new biological parts, devices, and systems or to redesign existing systems found in nature.

Condensins are large protein complexes that play a central role in chromosome assembly and segregation during mitosis and meiosis. Their subunits were originally identified as major components of mitotic chromosomes assembled in Xenopus egg extracts.
Heteroplasmy is the presence of more than one type of organellar genome within a cell or individual. It is an important factor in considering the severity of mitochondrial diseases. Because most eukaryotic cells contain many hundreds of mitochondria with hundreds of copies of mitochondrial DNA, it is common for mutations to affect only some mitochondria, leaving most unaffected.

Komagataella is a methylotrophic yeast within the order Saccharomycetales. It was found in the 1960s as Pichia pastoris, with its feature of using methanol as a source of carbon and energy. In 1995, P. pastoris was reassigned into the sole representative of genus Komagataella, becoming Komagataella pastoris. Later studies have further distinguished new species in this genus, resulting in a total of 7 recognized species. It is not uncommon to see the old name still in use, as of 2023; in less formal use, the yeast may confusingly be referred to as pichia.
Oligodendrocyte progenitor cells (OPCs), also known as oligodendrocyte precursor cells, NG2-glia, O2A cells, or polydendrocytes, are a subtype of glia in the central nervous system named for their essential role as precursors to oligodendrocytes. They are typically identified in the human by co-expression of PDGFRA and CSPG4.
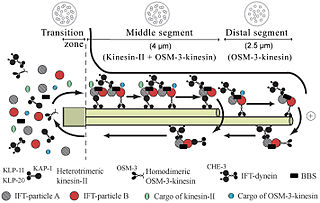
Intraflagellar transport (IFT) is a bidirectional motility along axoneme microtubules that is essential for the formation (ciliogenesis) and maintenance of most eukaryotic cilia and flagella. It is thought to be required to build all cilia that assemble within a membrane projection from the cell surface. Plasmodium falciparum cilia and the sperm flagella of Drosophila are examples of cilia that assemble in the cytoplasm and do not require IFT. The process of IFT involves movement of large protein complexes called IFT particles or trains from the cell body to the ciliary tip and followed by their return to the cell body. The outward or anterograde movement is powered by kinesin-2 while the inward or retrograde movement is powered by cytoplasmic dynein 2/1b. The IFT particles are composed of about 20 proteins organized in two subcomplexes called complex A and B.
Recombineering is a genetic and molecular biology technique based on homologous recombination systems, as opposed to the older/more common method of using restriction enzymes and ligases to combine DNA sequences in a specified order. Recombineering is widely used for bacterial genetics, in the generation of target vectors for making a conditional mouse knockout, and for modifying DNA of any source often contained on a bacterial artificial chromosome (BAC), among other applications.
Mycoplasma laboratorium or Synthia refers to a synthetic strain of bacterium. The project to build the new bacterium has evolved since its inception. Initially the goal was to identify a minimal set of genes that are required to sustain life from the genome of Mycoplasma genitalium, and rebuild these genes synthetically to create a "new" organism. Mycoplasma genitalium was originally chosen as the basis for this project because at the time it had the smallest number of genes of all organisms analyzed. Later, the focus switched to Mycoplasma mycoides and took a more trial-and-error approach.

Gail Roberta Martin is an American biologist. She is professor emerita in the Department of Anatomy, University of California, San Francisco. She is known for her pioneering work on the isolation of pluripotent stem cells from normal embryos, for which she coined the term ‘embryonic stem cells’. She is also widely recognized for her work on the function of Fibroblast Growth Factors (FGFs) and their negative regulators in vertebrate organogenesis. She and her colleagues also made valuable contributions to gene targeting technology.

A HEAT repeat is a protein tandem repeat structural motif composed of two alpha helices linked by a short loop. HEAT repeats can form alpha solenoids, a type of solenoid protein domain found in a number of cytoplasmic proteins. The name "HEAT" is an acronym for four proteins in which this repeat structure is found: Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1. HEAT repeats form extended superhelical structures which are often involved in intracellular transport; they are structurally related to armadillo repeats. The nuclear transport protein importin beta contains 19 HEAT repeats.

Genetically modified fish are organisms from the taxonomic clade which includes the classes Agnatha, Chondrichthyes and Osteichthyes whose genetic material (DNA) has been altered using genetic engineering techniques. In most cases, the aim is to introduce a new trait to the fish which does not occur naturally in the species, i.e. transgenesis.
Stuart C. Sealfon is an American neurologist who studies the mechanisms of both the therapeutic and adverse effects of drugs. He was an early adopter of the use of massively parallel qPCR and fluorescent in situ hybridization to characterize cell response state and his research accomplishments have included the identification of the primary structure of the gonadotropin-releasing hormone receptor, finding new signaling pathways activated by drugs for Parkinson's disease, elucidating the mechanism of action of hallucinogens and finding a new brain receptor complex implicated in schizophrenia as a novel target for antipsychotics.
Michael B. Elowitz is a biologist and professor of Biology, Bioengineering, and Applied Physics at the California Institute of Technology, and investigator at the Howard Hughes Medical Institute. In 2007 he was the recipient of the Genius grant, better known as the MacArthur Fellows Program for the design of a synthetic gene regulatory network, the Repressilator, which helped initiate the field of synthetic biology. He was the first to show how inherently random effects, or 'noise', in gene expression could be detected and quantified in living cells, leading to a growing recognition of the many roles that noise plays in living cells. His work in Synthetic Biology and Noise represent two foundations of the field of Systems Biology. Since then, his laboratory has contributed to the development of synthetic biological circuits that perform a range of functions inside cells, and revealed biological circuit design principles underlying epigenetic memory, cell fate control, cell-cell communication, and multicellular behaviors.

Synthetic biological circuits are an application of synthetic biology where biological parts inside a cell are designed to perform logical functions mimicking those observed in electronic circuits. The applications range from simply inducing production to adding a measurable element, like GFP, to an existing natural biological circuit, to implementing completely new systems of many parts.
Bovine seminal RNase (BS-RNase) is a member of the ribonuclease superfamily produced by the bovine seminal vesicles. This enzyme can not be differentiated from its members distinctly since there are more features that this enzyme shares with its family members than features that it possess alone. The research on the question of how new functions arrive in proteins in evolution led the scientists to find an uncommon consequence for a usual biological event called gene conversion in the case of the ribonuclease (RNase) protein family. The most well-known member of this family, RNase A, is expressed in the pancreas of oxen. It serves to digest RNA in intestine, and evolved from bacteria fermenting in the stomach of the first ox. The homologous RNase, called seminal RNase, differs from RNase A by 23 amino acids and is expressed in seminal plasma in the concentration of 1-1.5 mg/ml, which constitutes more than 3% of the fluid protein content. Bovine seminal ribonuclease (BS-RNase) is a homologue of RNase A with specific antitumor activity.
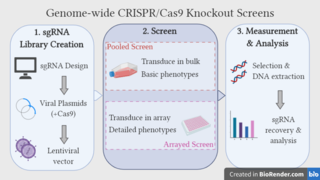
Genome-wide CRISPR-Cas9 knockout screens aim to elucidate the relationship between genotype and phenotype by ablating gene expression on a genome-wide scale and studying the resulting phenotypic alterations. The approach utilises the CRISPR-Cas9 gene editing system, coupled with libraries of single guide RNAs (sgRNAs), which are designed to target every gene in the genome. Over recent years, the genome-wide CRISPR screen has emerged as a powerful tool for performing large-scale loss-of-function screens, with low noise, high knockout efficiency and minimal off-target effects.
Xenopus egg extract is a lysate that is prepared by crushing the eggs of the African clawed frog Xenopus laevis. It offers a powerful cell-free system for studying various cell biological processes, including cell cycle progression, nuclear transport, DNA replication and chromosome segregation. It is also called Xenopus egg cell-free system or Xenopus egg cell-free extract.











