
Arachidonic acid is a polyunsaturated omega-6 fatty acid 20:4(ω-6), or 20:4(5,8,11,14). It is structurally related to the saturated arachidic acid found in cupuaçu butter. Its name derives from the Neo-Latin word arachis (peanut), but peanut oil does not contain any arachidonic acid.
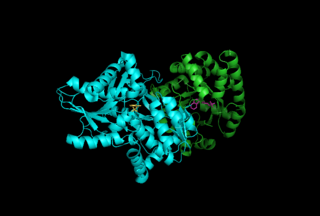
Tryptophan synthase or tryptophan synthetase is an enzyme that catalyses the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 angstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled.

Lipoxygenases are a family of (non-heme) iron-containing enzymes most of which catalyze the dioxygenation of polyunsaturated fatty acids in lipids containing a cis,cis-1,4- pentadiene into cell signaling agents that serve diverse roles as autocrine signals that regulate the function of their parent cells, paracrine signals that regulate the function of nearby cells, and endocrine signals that regulate the function of distant cells.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction
Arachidonate 5-lipoxygenase, also known as ALOX5, 5-lipoxygenase, 5-LOX, or 5-LO, is a non-heme iron-containing enzyme that in humans is encoded by the ALOX5 gene. Arachidonate 5-lipoxygenase is a member of the lipoxygenase family of enzymes. It transforms essential fatty acids (EFA) substrates into leukotrienes as well as a wide range of other biologically active products. ALOX5 is a current target for pharmaceutical intervention in a number of diseases.

Mead acid is an omega-9 fatty acid, first characterized by James F. Mead. As with some other omega-9 polyunsaturated fatty acids, animals can make Mead acid de novo. Its elevated presence in the blood is an indication of essential fatty acid deficiency. Mead acid is found in large quantities in cartilage.

12-oxophytodienoate reductase (OPRs) is an enzyme of the family of Old Yellow Enzymes (OYE). OPRs are grouped into two groups: OPRI and OPRII – the second group is the focus of this article, as the function of the first group is unknown, but is the subject of current research. The OPR enzyme utilizes the cofactor flavin mononucleotide (FMN) and catalyzes the following reaction in the jasmonic acid synthesis pathway:

ALOX15 is, like other lipoxygenases, a seminal enzyme in the metabolism of polyunsaturated fatty acids to a wide range of physiologically and pathologically important products. ▼ Gene Function

ALOX12, also known as arachidonate 12-lipoxygenase, 12-lipoxygenase, 12S-Lipoxygenase, 12-LOX, and 12S-LOX is a lipoxygenase-type enzyme that in humans is encoded by the ALOX12 gene which is located along with other lipoyxgenases on chromosome 17p13.3. ALOX12 is 75 kilodalton protein composed of 663 amino acids.
In enzymology, a linoleate 11-lipoxygenase (EC 1.13.11.45) is an enzyme that catalyzes the chemical reaction
In enzymology, a linoleate diol synthase (EC 1.13.11.44) is an enzyme that catalyzes the chemical reaction

Cyclooxygenase 1 (COX-1), also known as prostaglandin G/H synthase 1, prostaglandin-endoperoxide synthase 1 or prostaglandin H2 synthase 1, is an enzyme that in humans is encoded by the PTGS1 gene. In humans it is one of two cyclooxygenases.

The enzyme cutinase is a member of the hydrolase family. It catalyzes the following reaction:

Arachidonate 12-lipoxygenase, 12R type, also known as ALOX12B, 12R-LOX, and arachidonate lipoxygenase 3, is a lipoxygenase-type enzyme composed of 701 amino acids and encoded by the ALOX12B gene. The gene is located on chromosome 17 at position 13.1 where it forms a cluster with two other lipoxygenases, ALOXE3 and ALOX15B. Among the human lipoxygenases, ALOX12B is most closely related in amino acid sequence to ALOXE3

Epidermis-type lipoxygenase 3 is a member of the lipoxygenase family of enzymes; in humans, it is encoded by the ALOXE3 gene. This gene is located on chromosome 17 at position 13.1 where it forms a cluster with two other lipoxygenases, ALOX12B and ALOX15B. Among the human lipoxygenases, ALOXE3 is most closely related in amino acid sequence to ALOX12B. ALOXE3, ALOX12B, and ALOX15B are often classified as epidermal lipoxygenases, in distinction to the other three human lipoxygenases, because they were initially defined as being highly or even exclusively expressed and functioning in skin. The epidermis-type lipoxygenases are now regarded as a distinct subclass within the multigene family of mammalian lipoxygenases with mouse Aloxe3 being the ortholog to human ALOXE3, mouse Alox12b being the ortholog to human ALOX12B, and mouse Alox8 being the ortholog to human ALOX15B [supplied by OMIM]. ALOX12B and ALOXE3 in humans, Alox12b and Aloxe3 in mice, and comparable orthologs in other in other species are proposed to act sequentially in a multistep metabolic pathway that forms products that are structurally critical for creating and maintaining the skin's water barrier function.
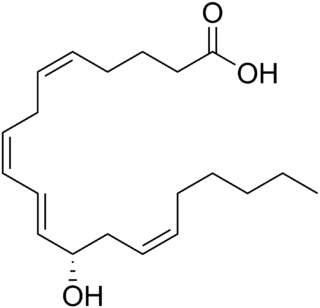
12-Hydroxyeicosatetraenoic acid (12-HETE) is a derivative of the 20 carbon polyunsaturated fatty acid, arachidonic acid, containing a hydroxyl residue at carbon 12 and a 5Z,8Z,10E,14Z Cis–trans isomerism configuration (Z=cis, E=trans) in its four double bonds. It was first found as a product of arachidonic acid metabolism made by human and bovine platelets through their 12S-lipoxygenase (i.e. ALOX12) enzyme(s). However, the term 12-HETE is ambiguous in that it has been used to indicate not only the initially detected "S" stereoisomer, 12S-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (12(S)-HETE or 12S-HETE), made by platelets, but also the later detected "R" stereoisomer, 12(R)-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (also termed 12(R)-HETE or 12R-HETE) made by other tissues through their 12R-lipoxygenase enzyme, ALOX12B. The two isomers, either directly or after being further metabolized, have been suggested to be involved in a variety of human physiological and pathological reactions. Unlike hormones which are secreted by cells, travel in the circulation to alter the behavior of distant cells, and thereby act as Endocrine signalling agents, these arachidonic acid metabolites act locally as Autocrine signalling and/or Paracrine signaling agents to regulate the behavior of their cells of origin or of nearby cells, respectively. In these roles, they may amplify or dampen, expand or contract cellular and tissue responses to disturbances.
Linoleate 8R-lipoxygenase (EC 1.13.11.60, linoleic acid 8R-dioxygenase, 5,8-LDS (bifunctional enzyme), 7,8-LDS (bifunctional enzyme), 5,8-linoleate diol synthase (bifunctional enzyme), 7,8-linoleate diol synthase (bifunctional enzyme), PpoA) is an enzyme with systematic name linoleate:oxygen (8R)-oxidoreductase. This enzyme catalyses the following chemical reaction
Linoleate 10R-lipoxygenase (EC 1.13.11.62, 10R-DOX, (10R)-dioxygenase, 10R-dioxygenase) is an enzyme with systematic name linoleate:oxygen (10R)-oxidoreductase. This enzyme catalyses the following chemical reaction
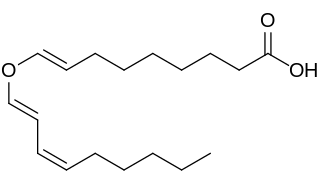
Divinylether fatty acids contain a fatty acid chemically combined with a doubly unsaturated carbon chain linked by an oxygen atom (ether). Fatty acid hydroperoxides generated by plant lipoxygenases from linoleic and linolenic acids are known to serve as substrates for a divinyl ether synthase which produces divinyl ether fatty acids. Up to date divinyl ethers were detected only within the plant kingdom.
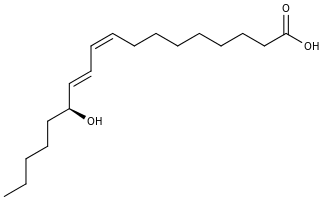
13-Hydroxyoctadecadienoic acid (13-HODE) is the commonly used term for 13(S)-hydroxy-9Z,11E-octadecadienoic acid (13(S)-HODE). The production of 13(S)-HODE is often accompanied by the production of its stereoisomer, 13(R)-hydroxy-9Z,11E-octadecadienoic acid (13(R)-HODE). The adjacent figure gives the structure for the (S) stereoisomer of 13-HODE. Two other naturally occurring 13-HODEs that may accompany the production of 13(S)-HODE are its cis-trans (i.e., 9E,11E) isomers viz., 13(S)-hydroxy-9E,11E-octadecadienoic acid (13(S)-EE-HODE) and 13(R)-hydroxy-9E,11E-octadecadienoic acid (13(R)-EE-HODE). Studies credit 13(S)-HODE with a range of clinically relevant bioactivities; recent studies have assigned activities to 13(R)-HODE that differ from those of 13(S)-HODE; and other studies have proposed that one or more of these HODEs mediate physiological and pathological responses, are markers of various human diseases, and/or contribute to the progression of certain diseases in humans. Since, however, many studies on the identification, quantification, and actions of 13(S)-HODE in cells and tissues have employed methods that did not distinguish between these isomers, 13-HODE is used here when the actual isomer studied is unclear.













