
Lactase is an enzyme produced by many organisms and is essential to the complete digestion of whole milk. It breaks down the sugar lactose into its component parts, galactose and glucose. Lactase is found in the brush border of the small intestine of humans and other mammals. People deficient in lactase or lacking functional lactase may experience the symptoms of lactose intolerance after consuming milk products. Microbial β-galactosidase can be purchased as a food supplement and is added to milk to produce "lactose-free" milk products.

Maltase is an informal name for a family of enzymes that catalyze the hydrolysis of disaccharide maltose into two simple sugars of glucose. Maltases are found in plants, bacteria, yeast, humans, and other vertebrates.

Acarbose (INN) is an anti-diabetic drug used to treat diabetes mellitus type 2 and, in some countries, prediabetes. It is a generic sold in Europe and China as Glucobay, in North America as Precose, and in Canada as Prandase.
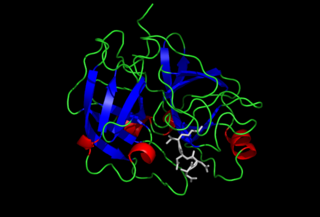
Enteropeptidase is an enzyme produced by cells of the duodenum and is involved in digestion in humans and other animals. Enteropeptidase converts trypsinogen into its active form trypsin, resulting in the subsequent activation of pancreatic digestive enzymes. Absence of enteropeptidase results in intestinal digestion impairment.

The glycogen debranching enzyme, in humans, is the protein encoded by the gene AGL. This enzyme is essential for the breakdown of glycogen, which serves as a store of glucose in the body. It has separate glucosyltransferase and glucosidase activities.
Isomaltase is an enzyme that breaks the bonds linking saccharides, which cannot be broken by amylase or maltase. It digests polysaccharides at the alpha 1-6 linkages. Its substrate, alpha-limit dextrin, is a product of amylopectin digestion that retains its 1-6 linkage. The product of the enzymatic digestion of alpha-limit dextrin by isomaltase is maltose.

1,4-alpha-glucan-branching enzyme, also known as brancher enzyme or glycogen-branching enzyme is an enzyme that in humans is encoded by the GBE1 gene.

Acid alpha-glucosidase, also called acid maltase, is an enzyme that helps to break down glycogen in the lysosome. It is functionally similar to glycogen debranching enzyme, but is on a different chromosome, processed differently by the cell and is located in the lysosome rather than the cytosol. In humans, it is encoded by the GAA gene. Errors in this gene cause glycogen storage disease type II.

α-Glucosidase (EC 3.2.1.20, is a glucosidase located in the brush border of the small intestine that acts upon α bonds:

The trefoil knot fold is a protein fold in which the protein backbone is twisted into a trefoil knot shape. "Shallow" knots in which the tail of the polypeptide chain only passes through a loop by a few residues are uncommon, but "deep" knots in which many residues are passed through the loop are extremely rare. Deep trefoil knots have been found in the SPOUT superfamily. including methyltransferase proteins involved in posttranscriptional RNA modification in all three domains of life, including bacterium Thermus thermophilus and proteins, in archaea and in eukaryota.

In biochemistry, glycoside hydrolases are a class of enzymes which catalyze the hydrolysis of glycosidic bonds in complex sugars. They are extremely common enzymes, with roles in nature including degradation of biomass such as cellulose (cellulase), hemicellulose, and starch (amylase), in anti-bacterial defense strategies, in pathogenesis mechanisms and in normal cellular function. Together with glycosyltransferases, glycosidases form the major catalytic machinery for the synthesis and breakage of glycosidic bonds.

Sucrose intolerance or genetic sucrase-isomaltase deficiency (GSID) is the condition in which sucrase-isomaltase, an enzyme needed for proper metabolism of sucrose (sugar) and starch, is not produced or the enzyme produced is either partially functional or non-functional in the small intestine. All GSID patients lack fully functional sucrase, while the isomaltase activity can vary from minimal functionality to almost normal activity. The presence of residual isomaltase activity may explain why some GSID patients are better able to tolerate starch in their diet than others with GSID.

Sucrase-isomaltase is a bifunctional glucosidase located on the brush border of the small intestine, encoded by the human gene SI. It is a dual-function enzyme with two GH31 domains, one serving as the isomaltase, the other as a sucrose alpha-glucosidase. It has preferential expression in the apical membranes of enterocytes. The enzyme’s purpose is to digest dietary carbohydrates such as starch, sucrose and isomaltose. By further processing the broken-down products, energy in the form of ATP can be generated.
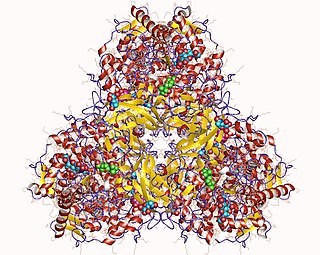
Glucan 1,4-α-glucosidase is an enzyme located on the brush border of the small intestine with systematic name 4-α-D-glucan glucohydrolase. It catalyses the following chemical reaction

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.

Glucosidase 2 subunit beta is an enzyme that in humans is encoded by the PRKCSH gene.
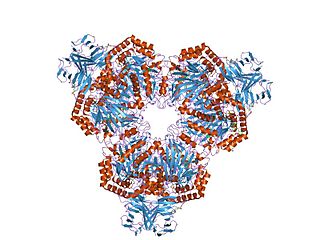
In molecular biology, glycoside hydrolase family 31 is a family of glycoside hydrolases.
In molecular biology, glycoside hydrolase family 15 is a family of glycoside hydrolases.
Limit dextrinase is an enzyme with systematic name dextrin 6-alpha-glucanohydrolase. This enzyme catalyses the hydrolysis of (1->6)-alpha-D-glucosidic linkages in alpha- and beta-limits dextrins of amylopectin and glycogen, in amylopectin and pullulan.
Sucrose α-glucosidase is an enzyme with systematic name sucrose-α-D-glucohydrolase. It catalyses the hydrolysis of sucrose and maltose by an α-D-glucosidase-type action.



















