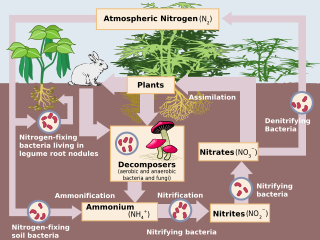
Denitrification is a microbially facilitated process where nitrate (NO3−) is reduced and ultimately produces molecular nitrogen (N2) through a series of intermediate gaseous nitrogen oxide products. Facultative anaerobic bacteria perform denitrification as a type of respiration that reduces oxidized forms of nitrogen in response to the oxidation of an electron donor such as organic matter. The preferred nitrogen electron acceptors in order of most to least thermodynamically favorable include nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O) finally resulting in the production of dinitrogen (N2) completing the nitrogen cycle. Denitrifying microbes require a very low oxygen concentration of less than 10%, as well as organic C for energy. Since denitrification can remove NO3−, reducing its leaching to groundwater, it can be strategically used to treat sewage or animal residues of high nitrogen content. Denitrification can leak N2O, which is an ozone-depleting substance and a greenhouse gas that can have a considerable influence on global warming.

Nitric oxide synthases (NOSs) are a family of enzymes catalyzing the production of nitric oxide (NO) from L-arginine. NO is an important cellular signaling molecule. It helps modulate vascular tone, insulin secretion, airway tone, and peristalsis, and is involved in angiogenesis and neural development. It may function as a retrograde neurotransmitter. Nitric oxide is mediated in mammals by the calcium-calmodulin controlled isoenzymes eNOS and nNOS. The inducible isoform, iNOS, involved in immune response, binds calmodulin at physiologically relevant concentrations, and produces NO as an immune defense mechanism, as NO is a free radical with an unpaired electron. It is the proximate cause of septic shock and may function in autoimmune disease.

Nitrosomonas is a genus of Gram-negative bacteria, belonging to the Betaproteobacteria. It is one of the five genera of ammonia-oxidizing bacteria and, as an obligate chemolithoautotroph, uses ammonia as an energy source and carbon dioxide as a carbon source in presence of oxygen. Nitrosomonas are important in the global biogeochemical nitrogen cycle, since they increase the bioavailability of nitrogen to plants and in the denitrification, which is important for the release of nitrous oxide, a powerful greenhouse gas. This microbe is photophobic, and usually generate a biofilm matrix, or form clumps with other microbes, to avoid light. Nitrosomonas can be divided into six lineages: the first one includes the species Nitrosomonas europea, Nitrosomonas eutropha, Nitrosomonas halophila, and Nitrosomonas mobilis. The second lineage presents the species Nitrosomonas communis, N. sp. I and N. sp. II, meanwhile the third lineage includes only Nitrosomonas nitrosa. The fourth lineage includes the species Nitrosomonas ureae and Nitrosomonas oligotropha and the fifth and sixth lineages include the species Nitrosomonas marina, N. sp. III, Nitrosomonas estuarii and Nitrosomonas cryotolerans.
Denitrifying bacteria are a diverse group of bacteria that encompass many different phyla. This group of bacteria, together with denitrifying fungi and archaea, is capable of performing denitrification as part of the nitrogen cycle. Denitrification is performed by a variety of denitrifying bacteria that are widely distributed in soils and sediments and that use oxidized nitrogen compounds in absence of oxygen as a terminal electron acceptor. They metabolise nitrogenous compounds using various enzymes, turning nitrogen oxides back to nitrogen gas or nitrous oxide.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Trimethylamine N-oxide reductase is a microbial enzyme that can reduce trimethylamine N-oxide (TMAO) into trimethylamine (TMA), as part of the electron transport chain. The enzyme has been purified from E. coli and the photosynthetic bacteria Roseobacter denitrificans.
In enzymology, a 4-hydroxybenzoyl-CoA reductase (EC 1.3.7.9) is an enzyme found in some bacteria and archaea that catalyzes the chemical reaction

In enzymology, a camphor 5-monooxygenase (EC 1.14.15.1) is an enzyme that catalyzes the chemical reaction

Nitric oxide dioxygenase (EC 1.14.12.17) is an enzyme that catalyzes the conversion of nitric oxide (NO) to nitrate (NO−
3) . The net reaction for the reaction catalyzed by nitric oxide dioxygenase is shown below:
In enzymology, a ferredoxin—nitrate reductase (EC 1.7.7.2) is an enzyme that catalyzes the chemical reaction
Hydroxylamine oxidoreductase (HAO) is an enzyme found in the prokaryotic genus Nitrosomonas. It plays a critically important role in the biogeochemical nitrogen cycle as part of the metabolism of ammonia-oxidizing bacteria.
In enzymology, a hydroxylamine reductase (EC 1.7.99.1) is an enzyme that catalyzes the chemical reaction
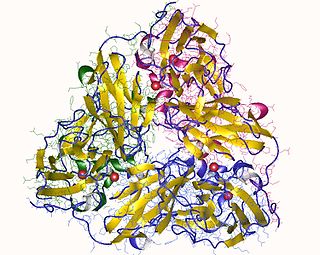
In enzymology, a nitrite reductase (NO-forming) (EC 1.7.2.1) is an enzyme that catalyzes the chemical reaction

In enzymology, a nitrous oxide reductase also known as nitrogen:acceptor oxidoreductase (N2O-forming) is an enzyme that catalyzes the final step in bacterial denitrification, the reduction of nitrous oxide to dinitrogen.
Aerobic denitrification or co-respiration the simultaneous use of both oxygen (O2) and nitrate (NO3−) as oxidizing agents, performed by various genera of microorganisms. This process differs from anaerobic denitrification not only in its insensitivity to the presence of oxygen, but also in that it has a higher potential to create the harmful byproduct nitrous oxide.
Nitric oxide reductase (NAD(P), nitrous oxide-forming) (EC 1.7.1.14, fungal nitric oxide reductase, cytochrome P450nor, NOR (ambiguous)) is an enzyme with systematic name nitrous oxide:NAD(P) oxidoreductase. This enzyme catalyses the following chemical reaction
Nitric oxide reductase (cytochrome c) (EC 1.7.2.5) is an enzyme with systematic name nitrous oxide:ferricytochrome-c oxidoreductase. This enzyme catalyses the following chemical reaction
Nitric oxide reductase (menaquinol) (EC 1.7.5.2) is an enzyme. This enzyme catalyses the following chemical reaction

Urine patches in cattle pastures generate large concentrations of the greenhouse gas nitrous oxide through nitrification and denitrification processes in urine-contaminated soils. Over the past few decades, the cattle population has increased more rapidly than the human population. Between the years 2000 and 2050, the cattle population is expected to increase from 1.5 billion to 2.6 billion. When large populations of cattle are packed into pastures, excessive amounts of urine soak into soils. This increases the rate at which nitrification and denitrification occur and produce nitrous oxide. Currently, nitrous oxide is one of the single most important ozone-depleting emissions and is expected to remain the largest throughout the 21st century.









