
Chymosin or rennin is a protease found in rennet. It is an aspartic endopeptidase belonging to MEROPS A1 family. It is produced by newborn ruminant animals in the lining of the abomasum to curdle the milk they ingest, allowing a longer residence in the bowels and better absorption. It is widely used in the production of cheese.
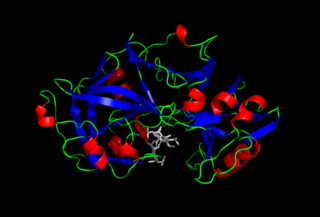
Pepsin is an endopeptidase that breaks down proteins into smaller peptides. It is produced in the gastric chief cells of the stomach lining and is one of the main digestive enzymes in the digestive systems of humans and many other animals, where it helps digest the proteins in food. Pepsin is an aspartic protease, using a catalytic aspartate in its active site.
In biochemistry, a zymogen, also called a proenzyme, is an inactive precursor of an enzyme. A zymogen requires a biochemical change for it to become an active enzyme. The biochemical change usually occurs in Golgi bodies, where a specific part of the precursor enzyme is cleaved in order to activate it. The inactivating piece which is cleaved off can be a peptide unit, or can be independently-folding domains comprising more than 100 residues. Although they limit the enzyme's ability, these N-terminal extensions of the enzyme or a “prosegment” often aid in the stabilization and folding of the enzyme they inhibit.
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.
Progastricsin also known as pepsinogen C or pepsinogen II is a pepsinogen precursor of the enzyme gastricsin that in humans is encoded by the PGC gene.

Cathepsin E is an enzyme that in humans is encoded by the CTSE gene. The enzyme is also known as slow-moving proteinase, erythrocyte membrane aspartic proteinase, SMP, EMAP, non-pepsin proteinase, cathepsin D-like acid proteinase, cathepsin E-like acid proteinase, cathepsin D-type proteinase) is an enzyme.
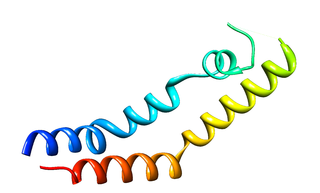
The plant-specific insert (PSI) or plant-specific sequence (PSS) is an independent domain, exclusively found in plants, consisting of approximately 100 residues, found on the C-terminal lobe on some aspartic proteases (AP) called phytepsins. The PSI, as an independent entity separate from its parent AP, is homologous to saposin and belongs to the saposin-like protein family (SAPLIP).
Penicillopepsin is an enzyme. This enzyme catalyses the following chemical reaction
Rhizopuspepsin is an enzyme. This enzyme catalyses the following chemical reaction
Endothiapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Candidapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Pycnoporopepsin is an enzyme. This enzyme catalyses the following chemical reaction

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.

Plasmepsin II (EC 3.4.23.39, aspartic hemoglobinase II, PFAPD) is an enzyme. This enzyme catalyses the following chemical reaction
Phytepsin is an enzyme. This enzyme catalyses the following chemical reaction
Leishmanolysin is an enzyme. This enzyme catalyses the following chemical reaction
Atrolysin C is an enzyme. This enzyme catalyses the following chemical reaction

The sedolisin family of peptidases are a family of serine proteases structurally related to the subtilisin (S8) family. Well-known members of this family include sedolisin ("pseudomonalisin") found in Pseudomonas bacteria, xanthomonalisin ("sedolisin-B"), physarolisin as well as animal tripeptidyl peptidase I. It is also known as sedolysin or serine-carboxyl peptidase. This group of enzymes contains a variation on the catalytic triad: unlike S8 which uses Ser-His-Asp, this group runs on Ser-Glu-Asp, with an additional acidic residue Asp in the oxyanion hole.









