
A bacteriophage, also known informally as a phage, is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.
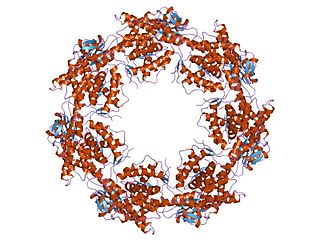
HSP60, also known as chaperonins (Cpn), is a family of heat shock proteins originally sorted by their 60kDa molecular mass. They prevent misfolding of proteins during stressful situations such as high heat, by assisting protein folding. HSP60 belong to a large class of molecules that assist protein folding, called molecular chaperones.

Filamentous bacteriophages are a family of viruses (Inoviridae) that infect bacteria, or bacteriophages. They are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. This distinctive shape reflects their method of replication: the coat of the virion comprises five types of viral protein, which are located in the inner membrane of the host bacterium during phage assembly, and these proteins are added to the nascent virion's DNA as it is extruded through the membrane. The simplicity of filamentous phages makes them an appealing model organism for research in molecular biology, and they have also shown promise as tools in nanotechnology and immunology.
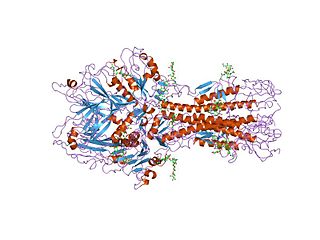
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as an invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C virus, toroviruses, and coronaviruses of the subgenus Embecovirus. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.
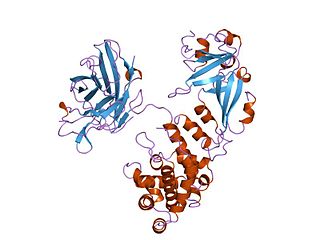
Diphtheria toxin is an exotoxin secreted mainly by Corynebacterium diphtheriae but also by Corynebacterium ulcerans and Corynebacterium pseudotuberculosis. the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.
Salmonella virus P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.

A DNA clamp, also known as a sliding clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.

Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

Phage typing is a phenotypic method that uses bacteriophages for detecting and identifying single strains of bacteria. Phages are viruses that infect bacteria and may lead to bacterial cell lysis. The bacterial strain is assigned a type based on its lysis pattern. Phage typing was used to trace the source of infectious outbreaks throughout the 1900s, but it has been replaced by genotypic methods such as whole genome sequencing for epidemiological characterization.

Streptococcal pyrogenic exotoxins also known as erythrogenic toxins, are exotoxins secreted by strains of the bacterial species Streptococcus pyogenes. SpeA and speC are superantigens, which induce inflammation by nonspecifically activating T cells and stimulating the production of inflammatory cytokines. SpeB, the most abundant streptococcal extracellular protein, is a cysteine protease. Pyrogenic exotoxins are implicated as the causative agent of scarlet fever and streptococcal toxic shock syndrome. There is no consensus on the exact number of pyrogenic exotoxins. Serotypes A-C are the most extensively studied and recognized by all sources, but others note up to thirteen distinct types, categorizing speF through speM as additional superantigens. Erythrogenic toxins are known to damage the plasma membranes of blood capillaries under the skin and produce a red skin rash. Past studies have shown that multiple variants of erythrogenic toxins may be produced, depending on the strain of S. pyogenes in question. Some strains may not produce a detectable toxin at all. Bacteriophage T12 infection of S. pyogenes enables the production of speA, and increases virulence.

In molecular biology, the Cro repressor family is a family of repressor proteins in bacteriophage lambda that includes the Cro repressor.

Ff phages is a group of almost identical filamentous phage including phages f1, fd, M13 and ZJ/2, which infect bacteria bearing the F fertility factor. The virion is a flexible filament measuring about 6 by 900 nm, comprising a cylindrical protein tube protecting a single-stranded circular DNA molecule at its core. The phage codes for only 11 gene products, and is one of the simplest viruses known. It has been widely used to study fundamental aspects of molecular biology. George Smith and Greg Winter used f1 and fd for their work on phage display for which they were awarded a share of the 2018 Nobel Prize in Chemistry. Early experiments on Ff phages used M13 to identify gene functions, and M13 was also developed as a cloning vehicle, so the name M13 is sometimes used as an informal synonym for the whole group of Ff phages.

cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host (lysis).
Phage-assisted continuous evolution (PACE) is a phage-based technique for the automated directed evolution of proteins. It relies on relating the desired activity of a target protein with the fitness of an infectious bacteriophage which carries the protein's corresponding gene. Proteins with greater desired activity hence confer greater infectivity to their carrier phage. More infectious phage propagate more effectively, selecting for advantageous mutations. Genetic variation is generated using error-prone polymerases on the phage vectors, and over time the protein accumulates beneficial mutations. This technique is notable for performing hundreds of rounds of selection with minimal human intervention.
















