Related Research Articles
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
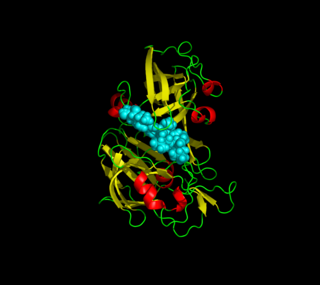
Plasmepsins are a class of at least 10 enzymes produced by the Plasmodium falciparum parasite. There are ten different isoforms of these proteins and ten genes coding them respectively in Plasmodium. It has been suggested that the plasmepsin family is smaller in other human Plasmodium species. Expression of Plm I, II, IV, V, IX, X and HAP occurs in the erythrocytic cycle, and expression of Plm VI, VII, VIII, occurs in the exoerythrocytic cycle. Through their haemoglobin-degrading activity, they are an important cause of symptoms in malaria sufferers. Consequently, this family of enzymes is a potential target for antimalarial drugs.

Pepsin A is an enzyme. This enzyme catalyses the following chemical reaction
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

In molecular biology, Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Parengyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.
Progastricsin also known as pepsinogen C or pepsinogen II is a pepsinogen precursor of the enzyme gastricsin that in humans is encoded by the PGC gene.
Nepenthesin is an aspartic protease of plant origin that has so far been identified in the pitcher secretions of Nepenthes and in the leaves of Drosera peltata. It is similar to pepsin, but differs in that it also cleaves on either side of Asp residues and at Lys┼Arg. While more pH and temperature stable than porcine pepsin A, it is considerably less stable in urea or guanidine hydrochloride. It is the only known protein with such a stability profile.
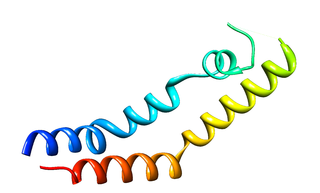
The plant-specific insert (PSI) or plant-specific sequence (PSS) is an independent domain, exclusively found in plants, consisting of approximately 100 residues, found on the C-terminal lobe on some aspartic proteases (AP) called phytepsins. The PSI, as an independent entity separate from its parent AP, is homologous to saposin and belongs to the saposin-like protein family (SAPLIP).

Aspergilloglutamic peptidase, also called aspergillopepsin II is a proteolytic enzyme. The enzyme was previously thought be an aspartic protease, but it was later shown to be a glutamic protease with a catalytic Glu residue at the active site, and was therefore renamed aspergilloglutamic peptidase.
Penicillopepsin is an enzyme. This enzyme catalyses the following chemical reaction
Endothiapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Mucorpepsin is an enzyme. This enzyme catalyses the following chemical reaction
Rhodotorulapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Acrocylindropepsin (EC 3.4.23.28, Acrocylindrium proteinase, Acrocylindrium acid proteinase) is an enzyme. This enzyme catalyses the following chemical reaction
Pycnoporopepsin is an enzyme. This enzyme catalyses the following chemical reaction
Scytalidopepsin A (EC 3.4.23.31, Scytalidium aspartic proteinase A, Scytalidium lignicolum aspartic proteinase, Scytalidium lignicolum aspartic proteinase A-2, Scytalidium lignicolum aspartic proteinase A-I, Scytalidium lignicolum aspartic proteinase C, Scytalidium lignicolum carboxyl proteinase, Scytalidium lignicolum acid proteinase) is an enzyme. This enzyme catalyses the following chemical reaction

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.
Deuterolysin is an enzyme. This enzyme catalyses the following chemical reaction

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.
References
- ↑ Tsuru D, Hattori A, Tsuji H, Yamamoto T, Fukumoto J (1969). "Studies on mold proteases. Part II. Substrate specificity of acid protease of Rhizopus chinensis". Agric. Biol. Chem. 33: 1419–1426. doi:10.1080/00021369.1969.10859482.
- ↑ Kurono, Y.; Chidimatsu, M.; Horikoshi, K.; Ikeda, Y. (1971). "Isolation of a protease from a Rhizopus product". Agric. Biol. Chem. 35 (11): 1668–1675. doi: 10.1271/bbb1961.35.1668 .
- ↑ Ohtsuru M, Tang J, Delaney R (1982). "Purification and characterization of rhizopuspepsin isozymes from a liquid culture of Rhizopus chinensis". The International Journal of Biochemistry. 14 (10): 925–32. doi:10.1016/0020-711x(82)90077-5. PMID 6751894.
- ↑ Suguna K, Padlan EA, Smith CW, Carlson WD, Davies DR (October 1987). "Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action". Proceedings of the National Academy of Sciences of the United States of America. 84 (20): 7009–13. doi: 10.1073/pnas.84.20.7009 . PMC 299218 . PMID 3313384.