Related Research Articles

In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.
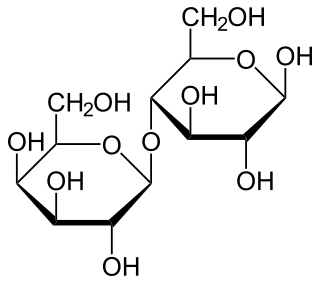
Lactose intolerance is caused by a lessened ability or a complete inability to digest lactose, a sugar found in dairy products. Humans vary in the amount of lactose they can tolerate before symptoms develop. Symptoms may include abdominal pain, bloating, diarrhea, flatulence, and nausea. These symptoms typically start thirty minutes to two hours after eating or drinking something containing lactose, with the severity typically depending on the amount consumed. Lactose intolerance does not cause damage to the gastrointestinal tract.

In genetics and bioinformatics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of considered population.
Researchers have investigated the relationship between race and genetics as part of efforts to understand how biology may or may not contribute to human racial categorization.
Lactase persistence is the continued activity of the lactase enzyme in adulthood, allowing the digestion of lactose in milk. In most mammals, the activity of the enzyme is dramatically reduced after weaning. In some human populations though, lactase persistence has recently evolved as an adaptation to the consumption of nonhuman milk and dairy products beyond infancy. Lactase persistence is very high among northern Europeans, especially Irish people. Worldwide, most people are lactase non-persistent, and are affected by varying degrees of lactose intolerance as adults. However, lactase persistence and lactose intolerance do not always overlap.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
In molecular biology, SNP array is a type of DNA microarray which is used to detect polymorphisms within a population. A single nucleotide polymorphism (SNP), a variation at a single site in DNA, is the most frequent type of variation in the genome. Around 335 million SNPs have been identified in the human genome, 15 million of which are present at frequencies of 1% or higher across different populations worldwide.
In genetics, a selective sweep is the process through which a new beneficial mutation that increases its frequency and becomes fixed in the population leads to the reduction or elimination of genetic variation among nucleotide sequences that are near the mutation. In selective sweep, positive selection causes the new mutation to reach fixation so quickly that linked alleles can "hitchhike" and also become fixed.
Human evolutionary genetics studies how one human genome differs from another human genome, the evolutionary past that gave rise to the human genome, and its current effects. Differences between genomes have anthropological, medical, historical and forensic implications and applications. Genetic data can provide important insights into human evolution.

The 1000 Genomes Project, launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one thousand anonymous participants from a number of different ethnic groups within the following three years, using newly developed technologies which were faster and less expensive. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal Nature. In 2012, the sequencing of 1092 genomes was announced in a Nature publication. In 2015, two papers in Nature reported results and the completion of the project and opportunities for future research.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.
Mark G. Thomas is a human evolutionary geneticist, Professor of Evolutionary Genetics at the Research Department of Genetics, Evolution and Environment at University College London since 2009. Prior to this, he was Cancer Research Campaign Postdoctoral Research Fellow at King's College London and then Wellcome Trust postdoctoral researcher in the department of Biological Anthropology at the University of Cambridge. He has acted as Editor-in-chief of the journal Annals of Human Genetics from 2015 to 2019 and Oct 2020 to Jan 2021.
Mitali Mukerji is a Professor and Head of the Department of Bioscience and Bioengineering, IIT Jodhpur. She was formerly a Chief Scientist at the CSIR Institute of Genomics and Integrative Biology with notable achievement in the field of human genomics and personalized medicine. She is best known for initiating the field of "Ayurgenomics" in partnership with her colleague Dr. Bhavana Prasher under the mentorship of Prof. Samir K. Brahmachari. Ayurgenomics is an innovative study, blending the principles of Ayurveda- the traditional Indian system of medicine- with genomics. Mukerji is also a major contributor in the Indian Genome Variation Consortium, a comprehensive database that is producing "the first genetic landscape of the Indian population", and has been an author in many publications that use IGV databases to study population genomics. Mukerji has done extensive research on hereditary ataxias, and is involved in many other projects related to tracking disease origins and mutational histories. She is the recipient of the prestigious Shanti Swarup Bhatnagar Award in 2010 for her contribution in the field of Medical Sciences.
Karin Broberg is a Swedish geneticist and toxicologist and professor at Karolinska Institutet and Lund University, Sweden, known for her work on human adaptation to challenging environments.
Recent human evolution refers to evolutionary adaptation, sexual and natural selection, and genetic drift within Homo sapiens populations, since their separation and dispersal in the Middle Paleolithic about 50,000 years ago. Contrary to popular belief, not only are humans still evolving, their evolution since the dawn of agriculture is faster than ever before. It has been proposed that human culture acts as a selective force in human evolution and has accelerated it; however, this is disputed. With a sufficiently large data set and modern research methods, scientists can study the changes in the frequency of an allele occurring in a tiny subset of the population over a single lifetime, the shortest meaningful time scale in evolution. Comparing a given gene with that of other species enables geneticists to determine whether it is rapidly evolving in humans alone. For example, while human DNA is on average 98% identical to chimp DNA, the so-called Human Accelerated Region 1 (HAR1), involved in the development of the brain, is only 85% similar.
Allele age is the amount of time elapsed since an allele first appeared due to mutation. Estimating the time at which a certain allele appeared allows researchers to infer patterns of human migration, disease, and natural selection. Allele age can be estimated based on (1) the frequency of the allele in a population and (2) the genetic variation that occurs within different copies of the allele, also known as intra-allelic variation. While either of these methods can be used to estimate allele age, the use of both increases the accuracy of the estimation and can sometimes offer additional information regarding the presence of selection.

Muntaser Eltayeb Ibrahim is a Sudanese geneticist and professor of molecular biology at the University of Khartoum, where he leads its Institute of Endemic Diseases. Science described him as "one of Sudan's most distinguished living scholars". His research focuses on human genetic diversity in Africa, human genetic variation contributing to susceptibility to infectious diseases such as malaria and leishmaniasis, and cancer genetics.

Charles Nohuoma Rotimi is the Scientific Director of the National Human Genome Research Institute (NHGRI). He joined the National Institutes of Health (NIH) in 2008 as the inaugural Director of the Trans-NIH Center for Research in Genomics and Global Health and was also the chief of the NHGRI's Metabolic, Cardiovascular, and Inflammatory Disease Genomics Branch. He works to ensure that population genetics include genomes from African populations and founded the African Society of Human Genetics in 2003 and was elected its first president. Rotimi was instrumental in the launch of the Human Heredity and Health in Africa (H3Africa) with the NIH and the Wellcome Trust. He was elected to the National Academy of Medicine in 2018.
Human genetic clustering refers to patterns of relative genetic similarity among human individuals and populations, as well as the wide range of scientific and statistical methods used to study this aspect of human genetic variation.

The African Society of Human Genetics (AfSHG) is a learned society and professional membership organization focused on the study of human genetics and genomics in Africans, and open to researchers who are interested in the subject. It has played a role in founding several national genetics societies, and is affiliated with the societies of Cameroon, the Democratic Republic of the Congo, Mali, Egypt, Rwanda, Senegal, South Africa, and Tanzania.
References
- 1 2 "Tishkoff Lab / Sarah Tishkoff, Ph.D." www.med.upenn.edu. Retrieved 2018-11-06.
- 1 2 3 4 5 Tishkoff, Sarah A. "Curriculum Vitae" (PDF). Retrieved 5 November 2018.
- 1 2 Tishkoff, S. A.; Dietzsch, E.; Speed, W.; Pakstis, A. J.; Kidd, J. R.; Cheung, K.; Bonné-Tamir, B.; Santachiara-Benerecetti, A. S.; Moral, P. (1996-03-08). "Global Patterns of Linkage Disequilibrium at the CD4 Locus and Modern Human Origins". Science. 271 (5254): 1380–1387. Bibcode:1996Sci...271.1380T. doi:10.1126/science.271.5254.1380. ISSN 0036-8075. PMID 8596909. S2CID 4266475.
- 1 2 3 4 Tishkoff, Sarah A.; Varkonyi, Robert; Cahinhinan, Nelie; Abbes, Salem; Argyropoulos, George; Destro-Bisol, Giovanni; Drousiotou, Anthi; Dangerfield, Bruce; Lefranc, Gerard (2001-07-20). "Haplotype Diversity and Linkage Disequilibrium at Human G6PD: Recent Origin of Alleles That Confer Malarial Resistance". Science. 293 (5529): 455–462. doi: 10.1126/science.1061573 . ISSN 0036-8075. PMID 11423617. S2CID 12230221.
- ↑ Tishkoff, Sarah A.; Reed, Floyd A.; Ranciaro, Alessia; Voight, Benjamin F.; Babbitt, Courtney C.; Silverman, Jesse S.; Powell, Kweli; Mortensen, Holly M.; Hirbo, Jibril B. (January 2007). "Convergent adaptation of human lactase persistence in Africa and Europe". Nature Genetics. 39 (1): 31–40. doi:10.1038/ng1946. ISSN 1061-4036. PMC 2672153 . PMID 17159977.
- ↑ Tishkoff, Sarah A.; Reed, Floyd A.; Friedlaender, Françoise R.; Ehret, Christopher; Ranciaro, Alessia; Froment, Alain; Hirbo, Jibril B.; Awomoyi, Agnes A.; Bodo, Jean-Marie (2009-05-22). "The Genetic Structure and History of Africans and African Americans". Science. 324 (5930): 1035–1044. Bibcode:2009Sci...324.1035T. doi:10.1126/science.1172257. ISSN 0036-8075. PMC 2947357 . PMID 19407144.
- ↑ Crawford, Nicholas G.; Kelly, Derek E.; Hansen, Matthew E. B.; Beltrame, Marcia H.; Fan, Shaohua; Bowman, Shanna L.; Jewett, Ethan; Ranciaro, Alessia; Thompson, Simon (2017-10-12). "Loci associated with skin pigmentation identified in African populations". Science. 358 (6365): eaan8433. doi:10.1126/science.aan8433. ISSN 0036-8075. PMC 5759959 . PMID 29025994.
- ↑ "National Academy of Sciences Elects Four Penn Professors | Penn Today". Penn Today. 5 May 2017. Retrieved 2018-11-06.
- 1 2 3 4 5 6 7 8 9 10 11 Tishkoff, S. A. (2018, November 2). Sarah Tishkoff - Early and Personal Life [Telephone interview].
- 1 2 3 "r/science - Science AMA Series: I study the population history and genetic diversity of Africa, human evolution, and the evolutionary dynamics of complex disease risk. I'm Sarah Tishkoff, a professor of genetics and biology at the UPenn School of Medicine, AMA!". reddit. Retrieved 2018-11-06.
- ↑ Tishkoff, Sarah. "LinkedIn" . Retrieved November 2, 2018.
- ↑ "Three Penn Medicine Scientists Elected to the National Academy of Sciences – PR News". www.pennmedicine.org. Retrieved 2018-11-06.
- 1 2 "Sarah Tishkoff". www.nasonline.org. Retrieved 2018-11-06.
- ↑ "Tishkoff Lab / Research". www.med.upenn.edu. Retrieved 2018-12-21.
- ↑ Beltrame, Marcia Holsbach; Rubel, Meagan A; Tishkoff, Sarah A (December 2016). "Inferences of African evolutionary history from genomic data". Current Opinion in Genetics & Development. 41: 159–166. doi:10.1016/j.gde.2016.10.002. ISSN 0959-437X. PMC 5161638 . PMID 27810637.
- ↑ Nielsen, Rasmus; Akey, Joshua M.; Jakobsson, Mattias; Pritchard, Jonathan K.; Tishkoff, Sarah; Willerslev, Eske (January 2017). "Tracing the peopling of the world through genomics". Nature. 541 (7637): 302–310. Bibcode:2017Natur.541..302N. doi:10.1038/nature21347. ISSN 0028-0836. PMC 5772775 . PMID 28102248.
- ↑ Hsieh, PingHsun; Woerner, August E.; Wall, Jeffrey D.; Lachance, Joseph; Tishkoff, Sarah A.; Gutenkunst, Ryan N.; Hammer, Michael F. (2016). "Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies". Genome Research. 26 (3): 291–300. doi:10.1101/gr.196634.115. ISSN 1088-9051. PMC 4772012 . PMID 26888264.
- ↑ Beltrame, Marcia Holsbach; Rubel, Meagan A; Tishkoff, Sarah A (2016). "Inferences of African evolutionary history from genomic data". Current Opinion in Genetics & Development. 41: 159–166. doi:10.1016/j.gde.2016.10.002. ISSN 0959-437X. PMC 5161638 . PMID 27810637.
- ↑ Campbell, Michael C.; Tishkoff, Sarah A. (2008). "African Genetic Diversity: Implications for Human Demographic History, Modern Human Origins, and Complex Disease Mapping". Annual Review of Genomics and Human Genetics. 9 (1): 403–433. doi:10.1146/annurev.genom.9.081307.164258. ISSN 1527-8204. PMC 2953791 . PMID 18593304.
- 1 2 Tishkoff, Sarah A.; Reed, Floyd A.; Friedlaender, Françoise R.; Ehret, Christopher; Ranciaro, Alessia; Froment, Alain; Hirbo, Jibril B.; Awomoyi, Agnes A.; Bodo, Jean-Marie (2009-05-22). "The Genetic Structure and History of Africans and African Americans". Science. 324 (5930): 1035–1044. Bibcode:2009Sci...324.1035T. doi:10.1126/science.1172257. ISSN 0036-8075. PMC 2947357 . PMID 19407144.
- 1 2 Campbell, Michael C; Hirbo, Jibril B; Townsend, Jeffrey P; Tishkoff, Sarah A (2014). "The peopling of the African continent and the diaspora into the new world". Current Opinion in Genetics & Development. 29: 120–132. doi:10.1016/j.gde.2014.09.003. ISSN 0959-437X. PMC 4308437 . PMID 25461616.
- 1 2 3 Gomez, Felicia; Tomas, Gil; Ko, Wen-Ya; Ranciaro, Alessia; Froment, Alain; Ibrahim, Muntaser; Lema, Godfrey; Nyambo, Thomas B.; Omar, Sabah A. (2013-04-23). "Patterns of nucleotide and haplotype diversity at ICAM-1 across global human populations with varying levels of malaria exposure". Human Genetics. 132 (9): 987–999. doi:10.1007/s00439-013-1284-5. ISSN 0340-6717. PMC 3916218 . PMID 23609612.
- 1 2 Ko, Wen-Ya; Rajan, Prianka; Gomez, Felicia; Scheinfeldt, Laura; An, Ping; Winkler, Cheryl A.; Froment, Alain; Nyambo, Thomas B.; Omar, Sabah A. (July 2013). "Identifying Darwinian Selection Acting on Different Human APOL1 Variants among Diverse African Populations". The American Journal of Human Genetics. 93 (1): 54–66. doi:10.1016/j.ajhg.2013.05.014. ISSN 0002-9297. PMC 3710747 . PMID 23768513.
- 1 2 3 Tishkoff, Sarah A; Reed, Floyd A; Ranciaro, Alessia; Voight, Benjamin F; Babbitt, Courtney C; Silverman, Jesse S; Powell, Kweli; Mortensen, Holly M; Hirbo, Jibril B (2006-12-10). "Convergent adaptation of human lactase persistence in Africa and Europe". Nature Genetics. 39 (1): 31–40. doi:10.1038/ng1946. ISSN 1061-4036. PMC 2672153 . PMID 17159977.
- ↑ Ranciaro, Alessia; Campbell, Michael C.; Hirbo, Jibril B.; Ko, Wen-Ya; Froment, Alain; Anagnostou, Paolo; Kotze, Maritha J.; Ibrahim, Muntaser; Nyambo, Thomas (April 2014). "Genetic Origins of Lactase Persistence and the Spread of Pastoralism in Africa". The American Journal of Human Genetics. 94 (4): 496–510. doi:10.1016/j.ajhg.2014.02.009. ISSN 0002-9297. PMC 3980415 . PMID 24630847.
- 1 2 3 Scheinfeldt, Laura B; Soi, Sameer; Thompson, Simon; Ranciaro, Alessia; Woldemeskel, Dawit; Beggs, William; Lambert, Charla; Jarvis, Joseph P; Abate, Dawit (2012). "Genetic adaptation to high altitude in the Ethiopian highlands". Genome Biology. 13 (1): R1. doi: 10.1186/gb-2012-13-1-r1 . ISSN 1465-6906. PMC 3334582 . PMID 22264333.
- 1 2 Campbell, M. C.; Ranciaro, A.; Froment, A.; Hirbo, J.; Omar, S.; Bodo, J.-M.; Nyambo, T.; Lema, G.; Zinshteyn, D. (2011-11-29). "Evolution of Functionally Diverse Alleles Associated with PTC Bitter Taste Sensitivity in Africa". Molecular Biology and Evolution. 29 (4): 1141–1153. doi:10.1093/molbev/msr293. ISSN 0737-4038. PMC 3341826 . PMID 22130969.
- ↑ Purba, Laurentia Henrieta Permita Sari; Widayati, Kanthi Arum; Tsutsui, Kei; Suzuki-Hashido, Nami; Hayakawa, Takashi; Nila, Sarah; Suryobroto, Bambang; Imai, Hiroo (2017-01-01). "Functional characterization of the TAS2R38 bitter taste receptor for phenylthiocarbamide in colobine monkeys". Biology Letters. 13 (1): 20160834. doi:10.1098/rsbl.2016.0834. ISSN 1744-9561. PMC 5310586 . PMID 28123110.
- 1 2 Jarvis, Joseph P.; Scheinfeldt, Laura B.; Soi, Sameer; Lambert, Charla; Omberg, Larsson; Ferwerda, Bart; Froment, Alain; Bodo, Jean-Marie; Beggs, William (2012-04-26). "Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies". PLOS Genetics. 8 (4): e1002641. doi: 10.1371/journal.pgen.1002641 . ISSN 1553-7404. PMC 3343053 . PMID 22570615.
- ↑ "Holiday Lectures | HHMI BioInteractive". www.hhmi.org. Retrieved 2018-12-21.
- ↑ "Sarah Tishkoff • iBiology". iBiology. Retrieved 2018-12-19.
- ↑ "About Us • iBiology". iBiology. Retrieved 2018-12-19.
- ↑ University of California Television (UCTV), CARTA: The Evolution of Human Biodiversity: Sarah Tishkoff - Local Adaptation , retrieved 2018-12-19
- ↑ "Burroughs Wellcome Fund" (PDF). Winter 2002. Retrieved November 5, 2018.
- ↑ "PIK :: Penn Integrates Knowledge – MEET THE PROFESSORS". pikprofessors.upenn.edu. Retrieved 2018-11-06.
- ↑ "Penn Geneticist Sarah A. Tishkoff Receives 2009 National Institutes of Health Pioneer Award | Penn Today". Penn Today. 24 September 2009. Retrieved 2018-11-06.
- ↑ "ASHG Honors Charles Rotimi & Sarah Tishkoff with 2019 Curt Stern Award | ASHG". www.ashg.org. 22 July 2019. Retrieved 2019-07-26.