
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

An ion source is a device that creates atomic and molecular ions. Ion sources are used to form ions for mass spectrometers, optical emission spectrometers, particle accelerators, ion implanters and ion engines.

Electrospray ionization (ESI) is a technique used in mass spectrometry to produce ions using an electrospray in which a high voltage is applied to a liquid to create an aerosol. It is especially useful in producing ions from macromolecules because it overcomes the propensity of these molecules to fragment when ionized. ESI is different from other ionization processes since it may produce multiple-charged ions, effectively extending the mass range of the analyser to accommodate the kDa-MDa orders of magnitude observed in proteins and their associated polypeptide fragments.

Koichi Tanaka is a Japanese electrical engineer who shared the Nobel Prize in Chemistry in 2002 for developing a novel method for mass spectrometric analyses of biological macromolecules with John Bennett Fenn and Kurt Wüthrich.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy-absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.

The history of mass spectrometry has its roots in physical and chemical studies regarding the nature of matter. The study of gas discharges in the mid 19th century led to the discovery of anode and cathode rays, which turned out to be positive ions and electrons. Improved capabilities in the separation of these positive ions enabled the discovery of stable isotopes of the elements. The first such discovery was with the element neon, which was shown by mass spectrometry to have at least two stable isotopes: 20Ne and 22Ne. Mass spectrometers were used in the Manhattan Project for the separation of isotopes of uranium necessary to create the atomic bomb.
Surface-enhanced laser desorption/ionization (SELDI) is a soft ionization method in mass spectrometry (MS) used for the analysis of protein mixtures. It is a variation of matrix-assisted laser desorption/ionization (MALDI). In MALDI, the sample is mixed with a matrix material and applied to a metal plate before irradiation by a laser, whereas in SELDI, proteins of interest in a sample become bound to a surface before MS analysis. The sample surface is a key component in the purification, desorption, and ionization of the sample. SELDI is typically used with time-of-flight (TOF) mass spectrometers and is used to detect proteins in tissue samples, blood, urine, or other clinical samples, however, SELDI technology can potentially be used in any application by simply modifying the sample surface.

Gentisic acid is a dihydroxybenzoic acid. It is a derivative of benzoic acid and a minor (1%) product of the metabolic break down of aspirin, excreted by the kidneys.

The compound 3-nitrobenzyl alcohol is an organic compound with the formula C7H7NO3.

MALDI mass spectrometry imaging (MALDI-MSI) is the use of matrix-assisted laser desorption ionization as a mass spectrometry imaging technique in which the sample, often a thin tissue section, is moved in two dimensions while the mass spectrum is recorded. Advantages, like measuring the distribution of a large amount of analytes at one time without destroying the sample, make it a useful method in tissue-based study.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.

Matrix-assisted laser desorption electrospray ionization (MALDESI) was first introduced in 2006 as a novel ambient ionization technique which combines the benefits of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI to resonantly excite an endogenous or exogenous matrix. The term 'matrix' refers to any molecule that is present in large excess and absorbs the energy of the laser, thus facilitating desorption of analyte molecules. The original MALDESI design was implemented using common organic matrices, similar to those used in MALDI, along with a UV laser. The current MALDESI source employs endogenous water or a thin layer of exogenously deposited ice as the energy-absorbing matrix where O-H symmetric and asymmetric stretching bonds are resonantly excited by a mid-IR laser.
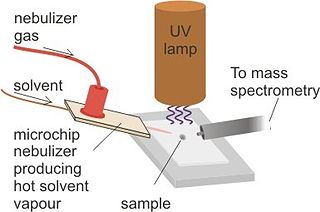
Desorption atmospheric pressure photoionization (DAPPI) is an ambient ionization technique for mass spectrometry that uses hot solvent vapor for desorption in conjunction with photoionization. Ambient Ionization techniques allow for direct analysis of samples without pretreatment. The direct analysis technique, such as DAPPI, eliminates the extraction steps seen in most nontraditional samples. DAPPI can be used to analyze bulkier samples, such as, tablets, powders, resins, plants, and tissues. The first step of this technique utilizes a jet of hot solvent vapor. The hot jet thermally desorbs the sample from a surface. The vaporized sample is then ionized by the vacuum ultraviolet light and consequently sampled into a mass spectrometer. DAPPI can detect a range of both polar and non-polar compounds, but is most sensitive when analyzing neutral or non-polar compounds. This technique also offers a selective and soft ionization for highly conjugated compounds.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.

Franz Hillenkamp was a German scientist known for his development of the laser microprobe mass analyzer and, with Michael Karas, matrix-assisted laser desorption/ionization (MALDI).
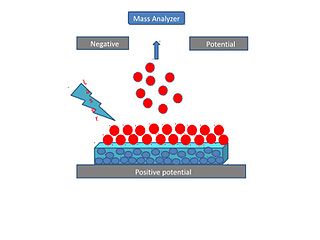
Surface-assisted laser desorption/ionization (SALDI) is a soft laser desorption technique used for mass spectrometry analysis of biomolecules, polymers, and small organic molecules. In its first embodiment Koichi Tanaka used a cobalt/glycerol liquid matrix and subsequent applications included a graphite/glycerol liquid matrix as well as a solid surface of porous silicon. The porous silicon represents the first matrix-free SALDI surface analysis allowing for facile detection of intact molecular ions, these porous silicon surfaces also facilitated the analysis of small molecules at the yoctomole level. At present laser desorption/ionization methods using other inorganic matrices such as nanomaterials are often regarded as SALDI variants. As an example, silicon nanowires as well as Titania nanotube arrays (NTA) have been used as substrates to detect small molecules. SALDI is used to detect proteins and protein-protein complexes. A related method named "ambient SALDI" - which is a combination of conventional SALDI with ambient mass spectrometry incorporating the direct analysis real time (DART) ion source has also been demonstrated. SALDI is considered one of the most important techniques in MS and has many applications.

Robert J. Cotter was an American chemist and mass spectrometrist. His research contributed to many early advances in the field of time-of-flight mass spectrometry. From 1998 to 2000 he was president of the American Society for Mass Spectrometry. Cotter was also a co-investigator on the Mars Organic Molecule Analyzer (MOMA) project, developing a miniaturized, low power consumption ion trap/time-of-flight mass spectrometer that was to be deployed with the ExoMars rover, now the Rosalind Franklin rover.

Desorption/ionization on silicon (DIOS) is a soft laser desorption method used to generate gas-phase ions for mass spectrometry analysis. DIOS is considered the first surface-based surface-assisted laser desorption/ionization (SALDI-MS) approach. Prior approaches were accomplished using nanoparticles in a matrix of glycerol, while DIOS is a matrix-free technique in which a sample is deposited on a nanostructured surface and the sample desorbed directly from the nanostructured surface through the adsorption of laser light energy. DIOS has been used to analyze organic molecules, metabolites, biomolecules and peptides, and, ultimately, to image tissues and cells.
In mass spectrometry, a matrix is a compound that promotes the formation of ions. Matrix compounds are used in matrix-assisted laser desorption/ionization (MALDI), matrix-assisted ionization (MAI), and fast atom bombardment (FAB).

















