
A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.

Stigmergy is a mechanism of indirect coordination, through the environment, between agents or actions. The principle is that the trace left in the environment by an individual action stimulates the performance of a succeeding action by the same or different agent. Agents that respond to traces in the environment receive positive fitness benefits, reinforcing the likelihood of these behaviors becoming fixed within a population over time.

The myxobacteria are a group of bacteria that predominantly live in the soil and feed on insoluble organic substances. The myxobacteria have very large genomes relative to other bacteria, e.g. 9–10 million nucleotides except for Anaeromyxobacter and Vulgatibacter. One species of myxobacteria, Minicystis rosea, has the largest known bacterial genome with over 16 million nucleotides. The second largest is another myxobacteria Sorangium cellulosum.

Myxococcus xanthus is a gram-negative, bacillus species of myxobacteria that exhibits various forms of self-organizing behavior in response to environmental cues. Under normal conditions with abundant food, it exists as a predatory, saprophytic single-species biofilm called a swarm. Under starvation conditions, it undergoes a multicellular development cycle.

Multicopy single-stranded DNA (msDNA) is a type of extrachromosomal satellite DNA that consists of a single-stranded DNA molecule covalently linked via a 2'-5'phosphodiester bond to an internal guanosine of an RNA molecule. The resultant DNA/RNA chimera possesses two stem-loops joined by a branch similar to the branches found in RNA splicing intermediates. The coding region for msDNA, called a "retron", also encodes a type of reverse transcriptase, which is essential for msDNA synthesis.
Sorangium cellulosum is a soil-dwelling Gram-negative bacterium of the group myxobacteria. It is motile and shows gliding motility. Under stressful conditions this motility, as in other myxobacteria, the cells congregate to form fruiting bodies and differentiate into myxospores. These congregating cells make isolation of pure culture and colony counts on agar medium difficult as the bacterium spread and colonies merge. It has an unusually-large genome of 13,033,779 base pairs, making it the largest bacterial genome sequenced to date by roughly 4 Mb.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.

Cruentarens are a group of macrolides secreted by the myxobacteria Byssovorax cruenta. There are two isomers (cruentaren A and B) have been isolated. They each have a molecular formula of C33H51NO8 and molecular weight 589 g/mol. Cruentaren A strongly inhibits the growth of yeasts and filamentous fungi, and inhibits the proliferation of different cancer cell lines in vitro, including a multidrug-resistant KB line. Cruentaren B shows only marginal cytotoxicity and no antifungal activity.

Gliding motility is a type of translocation used by microorganisms that is independent of propulsive structures such as flagella, pili, and fimbriae. Gliding allows microorganisms to travel along the surface of low aqueous films. The mechanisms of this motility are only partially known.
Diadenosine tetraphosphate or Ap4A is a putative alarmone, ubiquitous in nature being common to everything from bacteria to humans. It is made up of two adenosines joined together by a 5′-5′ linked chain of four phosphates. Adenosine polyphosphates are capable of inducing multiple physiological effects.
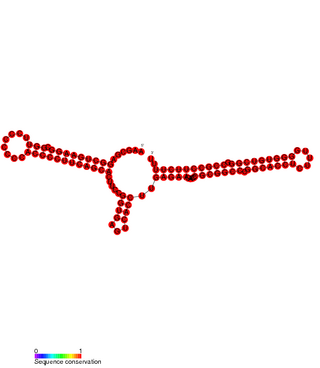
Pxr sRNA is a regulatory RNA which downregulates genes responsible for the formation of fruiting bodies in Myxococcus xanthus. Fruiting bodies are aggregations of myxobacteria formed when nutrients are scarce, the fruiting bodies permit a small number of the aggregated colony to transform into stress-resistant spores.
Armin Dale Kaiser was an American biochemist, molecular geneticist, molecular biologist and developmental biologist.

Myxococcus is a genus of bacteria in the family Myxococcaceae. Myxococci are Gram-negative, spore-forming, chemoorganotrophic, obligate aerobes. They are elongated rods with rounded or tapered ends, and they are nonflagellated. The cells move by gliding and can predate other bacteria. The genus has been isolated from soil.
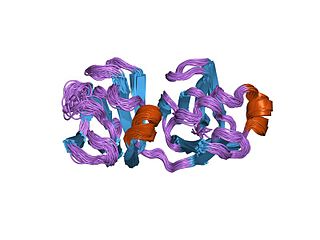
Protein S is a protein found in Myxococcus xanthus. Its name derives from being the "S" band in an alphabetical ordering of proteins run from Myxococcus xanthus cell contents on a SDS-denaturing gel. Its study was initially prompted by the huge increase in Protein S production during sporulation of Myxococcus xanthus.
Plesiocystis pacifica is a species of marine myxobacteria. Like other members of this order, P. pacifica is a rod-shaped Gram-negative bacterium that can move by gliding and can form aggregates of cells called fruiting bodies. The species was first described in 2003, based on two strains isolated from samples collected from the Pacific coast of Japan.
Enhygromyxa salina is a species of marine myxobacteria. Like other members of this order, E. salina is a rod-shaped Gram-negative bacterium that can move by gliding and can form aggregates of cells called fruiting bodies. E. salina is slightly halophilic (salt-tolerant) and can grow at lower temperatures than other marine myxobacteria. Several novel secondary metabolites have been identified in the species, including unusual sterols. The species was first described in 2003, based on six strains isolated from samples collected from the coastlines of Japan.

Social motility describes the motile movement of groups of cells that communicate with each other to coordinate movement based on external stimuli. There are multiple varieties of each kingdom that express social motility that provides a unique evolutionary advantages that other species do not possess. This has made them lethal killers such as African trypanosomiasis, or Myxobacteria. These evolutionary advantages have proven to increase survival rate among socially motile bacteria whether it be the ability to evade predators or communication within a swarm to form spores for long term hibernation in times of low nutrients or toxic environments.
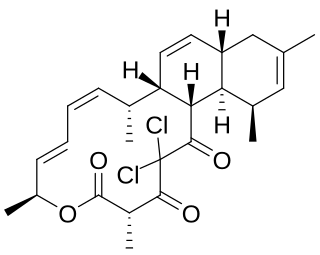
Chlorotonil A is a polyketide natural product produced by the myxobacterium Sorangium cellulosum So ce1525. It displays antimalarial activity in an animal model, and has in vitro antibacterial and antifungal activity. The activity of chlorotonil A has been attributed to the gem-dichloro-1,3-dione moiety, which is a unique functionality in polyketides. In addition to its unique halogenation, the structure of chlorotonil A has also garnered interest due to its similarity to anthracimycin, a polyketide natural product with antibiotic activity against Gram-positive bacteria.

Joshua Shaevitz is an American biophysicist and Professor of Physics at the Lewis-Sigler Institute at Princeton University in Princeton, NJ. He is known for his work in single-molecule biophysics, bacterial growth and motility, and animal behavior.

Adventurous motility is as a type of gliding motility; unlike most motility mechanisms, adventurous motility does not involve a flagellum. Gliding motility usually involves swarms of bacteria; however, adventurous motility is practiced by individual cells. This gliding is hypothesized to occur via assembly of a type IV secretion system and the extrusion of a polysaccharide slime, or by use of a series of adhesion complexes. The majority of research on adventurous motility has focused on the species, Myxococcus xanthus. The earliest of this research is attributed to Jonathan Hodgkin and Dale Kaiser.














