Related Research Articles
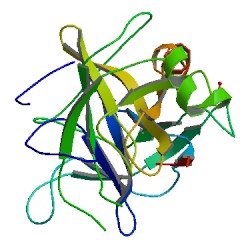
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.
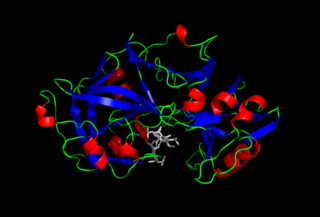
Pepsin is an endopeptidase that breaks down proteins into smaller peptides. It is produced in the gastric chief cells of the stomach lining and is one of the main digestive enzymes in the digestive systems of humans and many other animals, where it helps digest the proteins in food. Pepsin is an aspartic protease, using a catalytic aspartate in its active site.
An acid hydrolase is an enzyme that works best at acidic pHs. It is commonly located in lysosomes, which are acidic on the inside. Acid hydrolases may be nucleases, proteases, glycosidases, lipases, phosphatases, sulfatases and phospholipases and make up the approximately 50 degradative enzymes of the lysosome that break apart biological matter.
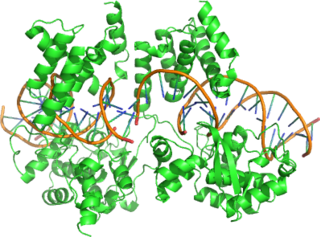
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the integrase family of site specific recombinase and it is known to catalyse the site specific recombination event between two DNA recognition sites. This 34 base pair (bp) loxP recognition site consists of two 13 bp palindromic sequences which flank an 8bp spacer region. The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. DNA sequences found between two loxP sites are said to be "floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites oriented in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are opposingly orientated will be inverted. The enzyme requires no additional cofactors or accessory proteins for its function.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.

Pepsin A is an enzyme. This enzyme catalyses the following chemical reaction

Nuclease S1 is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction
The discovery of an orally inactive peptide from snake venom established the important role of angiotensin converting enzyme (ACE) inhibitors in regulating blood pressure. This led to the development of captopril, the first ACE inhibitor. When the adverse effects of captopril became apparent new derivates were designed. Then after the discovery of two active sites of ACE: N-domain and C-domain, the development of domain-specific ACE inhibitors began.
In enzymology, a bis(5'-adenosyl)-triphosphatase (EC 3.6.1.29) is an enzyme that catalyzes the chemical reaction
In enzymology, a bis(5'-nucleosyl)-tetraphosphatase (asymmetrical) (EC 3.6.1.17) is an enzyme that catalyzes the chemical reaction
In enzymology, a bis(5'-nucleosyl)-tetraphosphatase (symmetrical) (EC 3.6.1.41) is an enzyme that catalyzes the chemical reaction
In enzymology, an ATP adenylyltransferase (EC 2.7.7.53) is an enzyme that catalyzes the chemical reaction
In enzymology, a guanosine-triphosphate guanylyltransferase (EC 2.7.7.45) is an enzyme that catalyzes the chemical reaction
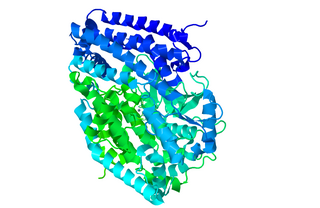
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.
Lysine carboxypeptidase is an enzyme. This enzyme catalyses the following chemical reaction:
Hepacivirin is an enzyme. This enzyme catalyses the following chemical reaction:
Streptopain is an enzyme. This enzyme catalyses the following chemical reaction
Gingipain R is an enzyme. This enzyme catalyses the following chemical reaction:
Mucorpepsin is an enzyme. This enzyme catalyses the following chemical reaction
Diadenosine hexaphosphate hydrolase (ATP-forming) (EC 3.6.1.61, Ndx1) is an enzyme with systematic name P1,P6-bis(5'-adenosyl)hexaphosphate nucleotidohydrolase (ATP-forming). This enzyme catalyses the following chemical reaction
References
- ↑ Lin X, Tang J (1995). Thermopsin. Methods in Enzymology. Vol. 248. pp. 156–68. doi:10.1016/0076-6879(95)48013-7. PMID 7674919.