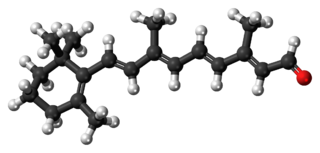
Retinal is a polyene chromophore. Retinal, bound to proteins called opsins, is the chemical basis of visual phototransduction, the light-detection stage of visual perception (vision).

Carotenoid oxygenases are a family of enzymes involved in the cleavage of carotenoids to produce, for example, retinol, commonly known as vitamin A. This family includes an enzyme known as RPE65 which is abundantly expressed in the retinal pigment epithelium where it catalyzed the formation of 11-cis-retinol from all-trans-retinyl esters.

In enzymology, an isovaleryl-CoA dehydrogenase is an enzyme that catalyzes the chemical reaction

In enzymology, beta-carotene 15,15'-dioxygenase, (EC 1.13.11.63) is an enzyme with systematic name beta-carotene:oxygen 15,15'-dioxygenase (bond-cleaving). In human it is encoded by the BCDO2 gene. This enzyme catalyses the following chemical reaction
In enzymology, tryptophan 2,3-dioxygenase (EC 1.13.11.11) is a heme enzyme that catalyzes the oxidation of L-tryptophan (L-Trp) to N-formyl-L-kynurenine, as the first and rate-limiting step of the kynurenine pathway.
In enzymology, a cytokinin dehydrogenase (EC 1.5.99.12) is an enzyme that catalyzes the chemical reaction
The enzyme chorismate synthase catalyzes the chemical reaction

Retinal pigment epithelium-specific 65 kDa protein, also known as retinoid isomerohydrolase, is an enzyme of the vertebrate visual cycle that is encoded in humans by the RPE65 gene. RPE65 is expressed in the retinal pigment epithelium and is responsible for the conversion of all-trans-retinyl esters to 11-cis-retinol during phototransduction. 11-cis-retinol is then used in visual pigment regeneration in photoreceptor cells. RPE65 belongs to the carotenoid oxygenase family of enzymes.
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine triphosphate (ATP) generation to xenobiotic degradation, the use of dioxygen as a biological oxidant is widespread and varied in the exact mechanism of its use. Enzymes employ many different schemes to use dioxygen, and this largely depends on the substrate and reaction at hand.
The one gene–one enzyme hypothesis is the idea that genes act through the production of enzymes, with each gene responsible for producing a single enzyme that in turn affects a single step in a metabolic pathway. The concept was proposed by George Beadle and Edward Tatum in an influential 1941 paper on genetic mutations in the mold Neurospora crassa, and subsequently was dubbed the "one gene–one enzyme hypothesis" by their collaborator Norman Horowitz. In 2004, Horowitz reminisced that "these experiments founded the science of what Beadle and Tatum called 'biochemical genetics.' In actuality they proved to be the opening gun in what became molecular genetics and all the developments that have followed from that." The development of the one gene–one enzyme hypothesis is often considered the first significant result in what came to be called molecular biology. Although it has been extremely influential, the hypothesis was recognized soon after its proposal to be an oversimplification. Even the subsequent reformulation of the "one gene–one polypeptide" hypothesis is now considered too simple to describe the relationship between genes and proteins.
Beta-apo-4'-carotenal oxygenase (EC 1.2.1.82, beta-apo-4'-carotenal dehydrogenase, YLO-1, carD (gene)) is an enzyme with systematic name 4'-apo-beta,psi-carotenal:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction:
Phytoene desaturase (3,4-didehydrolycopene-forming) is an enzyme with systematic name 15-cis-phytoene:acceptor oxidoreductase (3,4-didehydrolycopene-forming). This enzyme catalyses the following chemical reaction

Phytoene desaturase (lycopene-forming) are enzymes found in archaea, bacteria and fungi that are involved in carotenoid biosynthesis. They catalyze the conversion of colorless 15-cis-phytoene into a bright red lycopene in a biochemical pathway called the poly-trans pathway. The same process in plants and cyanobacteria utilizes four separate enzymes in a poly-cis pathway.
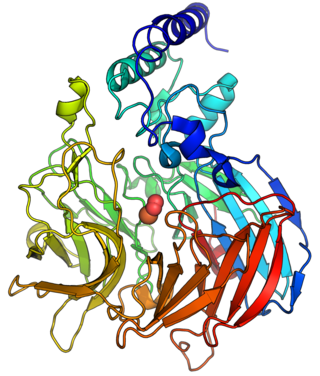
9-cis-epoxycarotenoid dioxygenase (EC 1.13.11.51, nine-cis-epoxycarotenoid dioxygenase, NCED, AtNCED3, PvNCED1, VP14) is an enzyme in the biosynthesis of abscisic acid (ABA), with systematic name 9-cis-epoxycarotenoid 11,12-dioxygenase. This enzyme catalyses the following chemical reaction
9-cis-beta-carotene 9',10'-cleaving dioxygenase (EC 1.13.11.68, CCD7 (gene), MAX3 (gene), NCED7 (gene)) is an enzyme with systematic name 9-cis-beta-carotene:O2 oxidoreductase (9',10'-cleaving). This enzyme catalyses the following chemical reaction
Carlactone synthase (EC 1.13.11.69, CCD8 (gene), MAX4 (gene), NCED8 (gene)) is an enzyme with systematic name 9-cis-10'-apo-beta-carotenal:O2 oxidoreductase (14,15-cleaving, carlactone-forming). This enzyme catalyses the following chemical reaction
Carotenoid-9',10'-cleaving dioxygenase (EC 1.13.11.71, BCO2 (gene), beta-carotene 9',10'-monooxygenase (misleading)) is an enzyme with systematic name all-trans-beta-carotene:O2 oxidoreductase (9',10'-cleaving). This enzyme catalyses the following chemical reaction
All-trans-8'-apo-beta-carotenal 15,15'-oxygenase (EC 1.14.99.41, Diox1, ACO, 8'-apo-beta-carotenal 15,15'-oxygenase) is an enzyme with systematic name all-trans-8'-apo-beta-carotenal:oxygen 15,15'-oxidoreductase (bond-cleaving). This enzyme catalyses the following chemical reaction
Zeaxanthin 7,8-dioxygenase (EC 1.14.99.42, zeaxanthin 7,8(7',8')-cleavage dioxygenase, CsZCD) is an enzyme with systematic name zeaxanthin:oxygen oxidoreductase (7,8-cleaving). This enzyme catalyses the following chemical reaction
Acireductone synthase (EC number 3.1.3.77, E1, E-1 enolase-phosphatase) is an enzyme with systematic name 5-(methylsulfanyl)-2,3-dioxopentyl-phosphate phosphohydrolase (isomerizing). It catalyses the following reaction:










