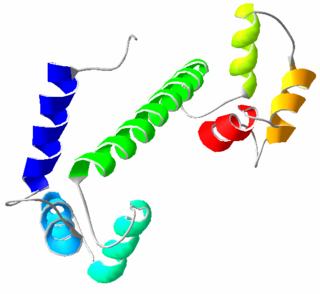
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.

Smooth muscle is an involuntary non-striated muscle, so-called because it has no sarcomeres and therefore no striations. It is divided into two subgroups, single-unit and multiunit smooth muscle. Within single-unit muscle, the whole bundle or sheet of smooth muscle cells contracts as a syncytium.

A myofibril is a basic rod-like organelle of a muscle cell. Skeletal muscles are composed of long, tubular cells known as muscle fibers, and these cells contain many chains of myofibrils. Each myofibril has a diameter of 1–2 micrometres. They are created during embryonic development in a process known as myogenesis.

A sarcomere is the smallest functional unit of striated muscle tissue. It is the repeating unit between two Z-lines. Skeletal muscles are composed of tubular muscle cells which are formed during embryonic myogenesis. Muscle fibers contain numerous tubular myofibrils. Myofibrils are composed of repeating sections of sarcomeres, which appear under the microscope as alternating dark and light bands. Sarcomeres are composed of long, fibrous proteins as filaments that slide past each other when a muscle contracts or relaxes. The costamere is a different component that connects the sarcomere to the sarcolemma.
Myosins are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.

Muscle contraction is the activation of tension-generating sites within muscle cells. In physiology, muscle contraction does not necessarily mean muscle shortening because muscle tension can be produced without changes in muscle length, such as when holding something heavy in the same position. The termination of muscle contraction is followed by muscle relaxation, which is a return of the muscle fibers to their low tension-generating state.
Voltage-gated calcium channels (VGCCs), also known as voltage-dependent calcium channels (VDCCs), are a group of voltage-gated ion channels found in the membrane of excitable cells (e.g., muscle, glial cells, neurons, etc.) with a permeability to the calcium ion Ca2+. These channels are slightly permeable to sodium ions, so they are also called Ca2+–Na+ channels, but their permeability to calcium is about 1000-fold greater than to sodium under normal physiological conditions.
CAMK, also written as CaMK or CCaMK, is an abbreviation for the Ca2+/calmodulin-dependent protein kinase class of enzymes. CAMKs are activated by increases in the concentration of intracellular calcium ions (Ca2+) and calmodulin. When activated, the enzymes transfer phosphates from ATP to defined serine or threonine residues in other proteins, so they are serine/threonine-specific protein kinases. Activated CAMK is involved in the phosphorylation of transcription factors and therefore, in the regulation of expression of responding genes. CAMK also works to regulate the cell life cycle (i.e. programmed cell death), rearrangement of the cell's cytoskeletal network, and mechanisms involved in the learning and memory of an organism.

Myofilaments are the three protein filaments of myofibrils in muscle cells. The main proteins involved are myosin, actin, and titin. Myosin and actin are the contractile proteins and titin is an elastic protein. The myofilaments act together in muscle contraction, and in order of size are a thick one of mostly myosin, a thin one of mostly actin, and a very thin one of mostly titin.

Telokin is an abundant protein found in smooth-muscle. It is identical to the C-terminus of myosin light-chain kinase. Telokin may play a role in the stabilization of unphosphorylated smooth-muscle myosin filaments. Because of its origin as the C-terminal end of smooth muscle myosin light chain kinase, it is called "telokin".
Calmodulin-binding proteins are, as their name implies, proteins which bind calmodulin. Calmodulin can bind to a variety of proteins through a two-step binding mechanism, namely "conformational and mutually induced fit", where typically two domains of calmodulin wrap around an emerging helical calmodulin binding domain from the target protein.

Caldesmon is a protein that in humans is encoded by the CALD1 gene.

Myosin light-chain phosphatase, also called myosin phosphatase (EC 3.1.3.53; systematic name [myosin-light-chain]-phosphate phosphohydrolase), is an enzyme (specifically a serine/threonine-specific protein phosphatase) that dephosphorylates the regulatory light chain of myosin II:

A myosin light chain is a light chain of myosin. Myosin light chains were discovered by Chinese biochemist Cao Tianqin when he was a graduate student at the University of Cambridge in England.

Calcium/calmodulin-dependent protein kinase type II gamma chain is an enzyme that in humans is encoded by the CAMK2G gene.

Myosin light chain kinase, smooth muscle also known as kinase-related protein (KRP) or telokin is an enzyme that in humans is encoded by the MYLK gene.

Myosin light chain kinase 3 also known as MYLK3, is an enzyme which in humans is encoded by the MYLK3 gene.

Myosin light chain kinase 2 also known as MYLK2 is an enzyme which in humans is encoded by the MYLK2 gene.

Rho-associated protein kinase (ROCK) is a kinase belonging to the AGC family of serine-threonine specific protein kinases. It is involved mainly in regulating the shape and movement of cells by acting on the cytoskeleton.

Calponin 1 is a basic smooth muscle protein that in humans is encoded by the CNN1 gene.


















