An action potential occurs when the membrane potential of a specific cell rapidly rises and falls. This depolarization then causes adjacent locations to similarly depolarize. Action potentials occur in several types of animal cells, called excitable cells, which include neurons, muscle cells, and in some plant cells. Certain endocrine cells such as pancreatic beta cells, and certain cells of the anterior pituitary gland are also excitable cells.
Computational neuroscience is a branch of neuroscience which employs mathematics, computer science, theoretical analysis and abstractions of the brain to understand the principles that govern the development, structure, physiology and cognitive abilities of the nervous system.
A Hopfield network is a spin glass system used to model neural networks, based on Ernst Ising's work with Wilhelm Lenz on the Ising model of magnetic materials. Hopfield networks were first described with respect to recurrent neural networks by Shun'ichi Amari in 1972 and with respect to biological neural networks by William Little in 1974, and were popularised by John Hopfield in 1982. Hopfield networks serve as content-addressable ("associative") memory systems with binary threshold nodes, or with continuous variables. Hopfield networks also provide a model for understanding human memory.

Neural oscillations, or brainwaves, are rhythmic or repetitive patterns of neural activity in the central nervous system. Neural tissue can generate oscillatory activity in many ways, driven either by mechanisms within individual neurons or by interactions between neurons. In individual neurons, oscillations can appear either as oscillations in membrane potential or as rhythmic patterns of action potentials, which then produce oscillatory activation of post-synaptic neurons. At the level of neural ensembles, synchronized activity of large numbers of neurons can give rise to macroscopic oscillations, which can be observed in an electroencephalogram. Oscillatory activity in groups of neurons generally arises from feedback connections between the neurons that result in the synchronization of their firing patterns. The interaction between neurons can give rise to oscillations at a different frequency than the firing frequency of individual neurons. A well-known example of macroscopic neural oscillations is alpha activity.
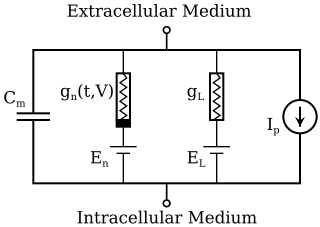
The Hodgkin–Huxley model, or conductance-based model, is a mathematical model that describes how action potentials in neurons are initiated and propagated. It is a set of nonlinear differential equations that approximates the electrical engineering characteristics of excitable cells such as neurons and muscle cells. It is a continuous-time dynamical system.

The FitzHugh–Nagumo model (FHN) describes a prototype of an excitable system.

Biological neuron models, also known as spiking neuron models, are mathematical descriptions of the conduction of electrical signals in neurons. Neurons are electrically excitable cells within the nervous system, able to fire electric signals, called action potentials, across a neural network.These mathematical models describe the role of the biophysical and geometrical characteristics of neurons on the conduction of electrical activity.
In neurophysiology, several mathematical models of the action potential have been developed, which fall into two basic types. The first type seeks to model the experimental data quantitatively, i.e., to reproduce the measurements of current and voltage exactly. The renowned Hodgkin–Huxley model of the axon from the Loligo squid exemplifies such models. Although qualitatively correct, the H-H model does not describe every type of excitable membrane accurately, since it considers only two ions, each with only one type of voltage-sensitive channel. However, other ions such as calcium may be important and there is a great diversity of channels for all ions. As an example, the cardiac action potential illustrates how differently shaped action potentials can be generated on membranes with voltage-sensitive calcium channels and different types of sodium/potassium channels. The second type of mathematical model is a simplification of the first type; the goal is not to reproduce the experimental data, but to understand qualitatively the role of action potentials in neural circuits. For such a purpose, detailed physiological models may be unnecessarily complicated and may obscure the "forest for the trees". The FitzHugh–Nagumo model is typical of this class, which is often studied for its entrainment behavior. Entrainment is commonly observed in nature, for example in the synchronized lighting of fireflies, which is coordinated by a burst of action potentials; entrainment can also be observed in individual neurons. Both types of models may be used to understand the behavior of small biological neural networks, such as the central pattern generators responsible for some automatic reflex actions. Such networks can generate a complex temporal pattern of action potentials that is used to coordinate muscular contractions, such as those involved in breathing or fast swimming to escape a predator.
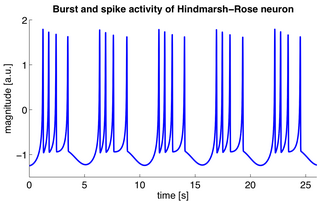
The Hindmarsh–Rose model of neuronal activity is aimed to study the spiking-bursting behavior of the membrane potential observed in experiments made with a single neuron. The relevant variable is the membrane potential, x(t), which is written in dimensionless units. There are two more variables, y(t) and z(t), which take into account the transport of ions across the membrane through the ion channels. The transport of sodium and potassium ions is made through fast ion channels and its rate is measured by y(t), which is called the spiking variable. z(t) corresponds to an adaptation current, which is incremented at every spike, leading to a decrease in the firing rate. Then, the Hindmarsh–Rose model has the mathematical form of a system of three nonlinear ordinary differential equations on the dimensionless dynamical variables x(t), y(t), and z(t). They read:
Models of neural computation are attempts to elucidate, in an abstract and mathematical fashion, the core principles that underlie information processing in biological nervous systems, or functional components thereof. This article aims to provide an overview of the most definitive models of neuro-biological computation as well as the tools commonly used to construct and analyze them.
The Morris–Lecar model is a biological neuron model developed by Catherine Morris and Harold Lecar to reproduce the variety of oscillatory behavior in relation to Ca++ and K+ conductance in the muscle fiber of the giant barnacle. Morris–Lecar neurons exhibit both class I and class II neuron excitability.
The network of the human nervous system is composed of nodes that are connected by links. The connectivity may be viewed anatomically, functionally, or electrophysiologically. These are presented in several Wikipedia articles that include Connectionism, Biological neural network, Artificial neural network, Computational neuroscience, as well as in several books by Ascoli, G. A. (2002), Sterratt, D., Graham, B., Gillies, A., & Willshaw, D. (2011), Gerstner, W., & Kistler, W. (2002), and Rumelhart, J. L., McClelland, J. L., and PDP Research Group (1986) among others. The focus of this article is a comprehensive view of modeling a neural network. Once an approach based on the perspective and connectivity is chosen, the models are developed at microscopic, mesoscopic, or macroscopic (system) levels. Computational modeling refers to models that are developed using computing tools.
Compartmental modelling of dendrites deals with multi-compartment modelling of the dendrites, to make the understanding of the electrical behavior of complex dendrites easier. Basically, compartmental modelling of dendrites is a very helpful tool to develop new biological neuron models. Dendrites are very important because they occupy the most membrane area in many of the neurons and give the neuron an ability to connect to thousands of other cells. Originally the dendrites were thought to have constant conductance and current but now it has been understood that they may have active Voltage-gated ion channels, which influences the firing properties of the neuron and also the response of neuron to synaptic inputs. Many mathematical models have been developed to understand the electric behavior of the dendrites. Dendrites tend to be very branchy and complex, so the compartmental approach to understand the electrical behavior of the dendrites makes it very useful.

The theta model, or Ermentrout–Kopell canonical model, is a biological neuron model originally developed to mathematically describe neurons in the animal Aplysia. The model is particularly well-suited to describe neural bursting, which is characterized by periodic transitions between rapid oscillations in the membrane potential followed by quiescence. This bursting behavior is often found in neurons responsible for controlling and maintaining steady rhythms such as breathing, swimming, and digesting. Of the three main classes of bursting neurons, the theta model describes parabolic bursting, which is characterized by a parabolic frequency curve during each burst.
In biology exponential integrate-and-fire models are compact and computationally efficient nonlinear spiking neuron models with one or two variables. The exponential integrate-and-fire model was first proposed as a one-dimensional model. The most prominent two-dimensional examples are the adaptive exponential integrate-and-fire model and the generalized exponential integrate-and-fire model. Exponential integrate-and-fire models are widely used in the field of computational neuroscience and spiking neural networks because of (i) a solid grounding of the neuron model in the field of experimental neuroscience, (ii) computational efficiency in simulations and hardware implementations, and (iii) mathematical transparency.
A binding neuron (BN) is an abstract concept of processing of input impulses in a generic neuron based on their temporal coherence and the level of neuronal inhibition. Mathematically, the concept may be implemented by most neuronal models including the well-known leaky integrate-and-fire model. The BN concept originated in 1996 and 1998 papers by A. K. Vidybida,
Dynamic causal modeling (DCM) is a framework for specifying models, fitting them to data and comparing their evidence using Bayesian model comparison. It uses nonlinear state-space models in continuous time, specified using stochastic or ordinary differential equations. DCM was initially developed for testing hypotheses about neural dynamics. In this setting, differential equations describe the interaction of neural populations, which directly or indirectly give rise to functional neuroimaging data e.g., functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG) or electroencephalography (EEG). Parameters in these models quantify the directed influences or effective connectivity among neuronal populations, which are estimated from the data using Bayesian statistical methods.
The spike response model (SRM) is a spiking neuron model in which spikes are generated by either a deterministic or a stochastic threshold process. In the SRM, the membrane voltage V is described as a linear sum of the postsynaptic potentials (PSPs) caused by spike arrivals to which the effects of refractoriness and adaptation are added. The threshold is either fixed or dynamic. In the latter case it increases after each spike. The SRM is flexible enough to account for a variety of neuronal firing pattern in response to step current input. The SRM has also been used in the theory of computation to quantify the capacity of spiking neural networks; and in the neurosciences to predict the subthreshold voltage and the firing times of cortical neurons during stimulation with a time-dependent current stimulation. The name Spike Response Model points to the property that the two important filters and of the model can be interpreted as the response of the membrane potential to an incoming spike (response kernel , the PSP) and to an outgoing spike (response kernel , also called refractory kernel). The SRM has been formulated in continuous time and in discrete time. The SRM can be viewed as a generalized linear model (GLM) or as an (integrated version of) a generalized integrate-and-fire model with adaptation.

Heteroclinic channels are ensembles of trajectories that can connect saddle equilibrium points in phase space. Dynamical systems and their associated phase spaces can be used to describe natural phenomena in mathematical terms; heteroclinic channels, and the cycles that they produce, are features in phase space that can be designed to occupy specific locations in that space. Heteroclinic channels move trajectories from one equilibrium point to another. More formally, a heteroclinic channel is a region in phase space in which nearby trajectories are drawn closer and closer to one unique limiting trajectory, the heteroclinic orbit. Equilibria connected by heteroclinic trajectories form heteroclinic cycles and cycles can be connected to form heteroclinic networks. Heteroclinic cycles and networks naturally appear in a number of applications, such as fluid dynamics, population dynamics, and neural dynamics. In addition, dynamical systems are often used as methods for robotic control. In particular, for robotic control, the equilibrium points can correspond to robotic states, and the heteroclinic channels can provide smooth methods for switching from state to state.

Synthetic Nervous System (SNS) is a computational neuroscience model that may be developed with the Functional Subnetwork Approach (FSA) to create biologically plausible models of circuits in a nervous system. The FSA enables the direct analytical tuning of dynamical networks that perform specific operations within the nervous system without the need for global optimization methods like genetic algorithms and reinforcement learning. The primary use case for a SNS is system control, where the system is most often a simulated biomechanical model or a physical robotic platform. An SNS is a form of a neural network much like artificial neural networks (ANNs), convolutional neural networks (CNN), and recurrent neural networks (RNN). The building blocks for each of these neural networks is a series of nodes and connections denoted as neurons and synapses. More conventional artificial neural networks rely on training phases where they use large data sets to form correlations and thus “learn” to identify a given object or pattern. When done properly this training results in systems that can produce a desired result, sometimes with impressive accuracy. However, the systems themselves are typically “black boxes” meaning there is no readily distinguishable mapping between structure and function of the network. This makes it difficult to alter the function, without simply starting over, or extract biological meaning except in specialized cases. The SNS method differentiates itself by using details of both structure and function of biological nervous systems. The neurons and synapse connections are intentionally designed rather than iteratively changed as part of a learning algorithm.