An autosome is any chromosome that is not a sex chromosome. The members of an autosome pair in a diploid cell have the same morphology, unlike those in allosomal pairs, which may have different structures. The DNA in autosomes is collectively known as atDNA or auDNA.

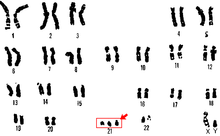
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

Aneuploidy is the presence of an abnormal number of chromosomes in a cell, for example a human cell having 45 or 47 chromosomes instead of the usual 46. It does not include a difference of one or more complete sets of chromosomes. A cell with any number of complete chromosome sets is called a euploid cell.

Cytogenetics is essentially a branch of genetics, but is also a part of cell biology/cytology, that is concerned with how the chromosomes relate to cell behaviour, particularly to their behaviour during mitosis and meiosis. Techniques used include karyotyping, analysis of G-banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH).

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division (mitosis/meiosis). There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously.

Prenatal testing is a tool that can be used to detect some birth defects at various stages prior to birth. Prenatal testing consists of prenatal screening and prenatal diagnosis, which are aspects of prenatal care that focus on detecting problems with the pregnancy as early as possible. These may be anatomic and physiologic problems with the health of the zygote, embryo, or fetus, either before gestation even starts or as early in gestation as practicable. Screening can detect problems such as neural tube defects, chromosome abnormalities, and gene mutations that would lead to genetic disorders and birth defects, such as spina bifida, cleft palate, Down syndrome, trisomy 18, Tay–Sachs disease, sickle cell anemia, thalassemia, cystic fibrosis, muscular dystrophy, and fragile X syndrome. Some tests are designed to discover problems which primarily affect the health of the mother, such as PAPP-A to detect pre-eclampsia or glucose tolerance tests to diagnose gestational diabetes. Screening can also detect anatomical defects such as hydrocephalus, anencephaly, heart defects, and amniotic band syndrome.
Comparative genomic hybridization (CGH) is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions. This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resolution of 5–10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.

A small supernumerary marker chromosome (sSMC) is an abnormal extra chromosome. It contains copies of parts of one or more normal chromosomes and like normal chromosomes is located in the cell's nucleus, is replicated and distributed into each daughter cell during cell division, and typically has genes which may be expressed. However, it may also be active in causing birth defects and neoplasms. The sSMC's small size makes it virtually undetectable using classical cytogenetic methods: the far larger DNA and gene content of the cell's normal chromosomes obscures those of the sSMC. Newer molecular techniques such as fluorescence in situ hybridization, next generation sequencing, comparative genomic hybridization, and highly specialized cytogenetic G banding analyses are required to study it. Using these methods, the DNA sequences and genes in sSMCs are identified and help define as well as explain any effect(s) it may have on individuals.
The Pallister–Killian syndrome (PKS), also termed tetrasomy 12p mosaicism or the Pallister mosaic aneuploidy syndrome, is an extremely rare and severe genetic disorder. PKS is due to the presence of an extra and abnormal chromosome termed a small supernumerary marker chromosome (sSMC). sSMCs contain copies of genetic material from parts of virtually any other chromosome and, depending on the genetic material they carry, can cause various genetic disorders and neoplasms. The sSMC in PKS consists of multiple copies of the short arm of chromosome 12. Consequently, the multiple copies of the genetic material in the sSMC plus the two copies of this genetic material in the two normal chromosome 12's are overexpressed and thereby cause the syndrome. Due to a form of genetic mosaicism, however, individuals with PKS differ in the tissue distributions of their sSMC and therefore show different syndrome-related birth defects and disease severities. For example, individuals with the sSMC in their heart tissue are likely to have cardiac structural abnormalities while those without this sSMC localization have a structurally normal heart.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.

Molecular cytogenetics combines two disciplines, molecular biology and cytogenetics, and involves the analysis of chromosome structure to help distinguish normal and cancer-causing cells. Human cytogenetics began in 1956 when it was discovered that normal human cells contain 46 chromosomes. However, the first microscopic observations of chromosomes were reported by Arnold, Flemming, and Hansemann in the late 1800s. Their work was ignored for decades until the actual chromosome number in humans was discovered as 46. In 1879, Arnold examined sarcoma and carcinoma cells having very large nuclei. Today, the study of molecular cytogenetics can be useful in diagnosing and treating various malignancies such as hematological malignancies, brain tumors, and other precursors of cancer. The field is overall focused on studying the evolution of chromosomes, more specifically the number, structure, function, and origin of chromosome abnormalities. It includes a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which DNA probes are labeled with different colored fluorescent tags to visualize one or more specific regions of the genome. Introduced in the 1980s, FISH uses probes with complementary base sequences to locate the presence or absence of the specific DNA regions. FISH can either be performed as a direct approach to metaphase chromosomes or interphase nuclei. Alternatively, an indirect approach can be taken in which the entire genome can be assessed for copy number changes using virtual karyotyping. Virtual karyotypes are generated from arrays made of thousands to millions of probes, and computational tools are used to recreate the genome in silico.
Confined placental mosaicism (CPM) represents a discrepancy between the chromosomal makeup of the cells in the placenta and the cells in the fetus. CPM was first described by Kalousek and Dill in 1983. CPM is diagnosed when some trisomic cells are detected on chorionic villus sampling and only normal cells are found on a subsequent prenatal test, such as amniocentesis or fetal blood sampling. In theory, CPM is when the trisomic cells are found only in the placenta. CPM is detected in approximately 1-2% of ongoing pregnancies that are studied by chorionic villus sampling (CVS) at 10 to 12 weeks of pregnancy. Chorionic villus sampling is a prenatal procedure which involves a placental biopsy. Most commonly when CPM is found it represents a trisomic cell line in the placenta and a normal diploid chromosome complement in the baby. However, the fetus is involved in about 10% of cases.

Not to be confused with Heterochromia, an optical condition which is commonly associated with cats
45,X/46,XY mosaicism, also known as X0/XY mosaicism and mixed gonadal dysgenesis, is a mutation of sex development in humans associated with sex chromosome aneuploidy and mosaicism of the Y chromosome. It is a fairly rare chromosomal disorder at birth, with an estimated incidence rate of about 1 in 15,000 live births. Mosaic loss of the Y chromosome in previously non-mosaic men grows increasingly common with age.

XYYY syndrome, also known as 48,XYYY, is a chromosomal disorder in which a male has two extra copies of the Y chromosome. The syndrome is exceptionally rare, with only twelve recorded cases. The presentation of the syndrome is heterogeneous, but appears to be more severe than its counterpart XYY syndrome. Common traits include borderline to mild intellectual disability, infertility, radioulnar synostosis, and in some cases tall stature.

Tetrasomy X, also known as 48,XXXX, is a chromosomal disorder in which a female has four, rather than two, copies of the X chromosome. It is associated with intellectual disability of varying severity, characteristic "coarse" facial features, heart defects, and skeletal anomalies such as increased height, clinodactyly, and radioulnar synostosis. Tetrasomy X is a rare condition, with few medically recognized cases; it is estimated to occur in approximately 1 in 50,000 females.

Pentasomy X, also known as 49,XXXXX, is a chromosomal disorder in which a female has five, rather than two, copies of the X chromosome. Pentasomy X is associated with short stature, intellectual disability, characteristic facial features, heart defects, skeletal anomalies, and pubertal and reproductive abnormalities. The condition is exceptionally rare, with an estimated prevalence between 1 in 85,000 and 1 in 250,000.

Trisomy X, also known as triple X syndrome and characterized by the karyotype 47,XXX, is a chromosome disorder in which a female has an extra copy of the X chromosome. It is relatively common and occurs in 1 in 1,000 females, but is rarely diagnosed; fewer than 10% of those with the condition know they have it.

XXXYY syndrome, also known as 49,XXXYY, is a chromosomal disorder in which a male has three copies of the X chromosome and two copies of the Y chromosome. XXXYY syndrome is exceptionally rare, with only eight recorded cases. Little is known about its presentation, but associated characteristics include intellectual disability, anomalies of the external genitalia, and characteristic physical and facial features. It is not caused by characteristics of the parents, but rather occurs via nondisjunction, a random event in gamete development. The karyotype observed in the syndrome is formally known as 49,XXXYY, which represents the 49 chromosomes observed in the disorder as compared to the 46 in normal human development.