Related Research Articles

Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions), and an imidazole side chain (which is partially protonated), classifying it as a positively charged amino acid at physiological pH. Initially thought essential only for infants, it has now been shown in longer-term studies to be essential for adults also. It is encoded by the codons CAU and CAC.

A hemeprotein, or heme protein, is a protein that contains a heme prosthetic group. They are a very large class of metalloproteins. The heme group confers functionality, which can include oxygen carrying, oxygen reduction, electron transfer, and other processes. Heme is bound to the protein either covalently or noncovalently or both.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.
Mothers against decapentaplegic homolog 2 also known as SMAD family member 2 or SMAD2 is a protein that in humans is encoded by the SMAD2 gene. MAD homolog 2 belongs to the SMAD, a family of proteins similar to the gene products of the Drosophila gene 'mothers against decapentaplegic' (Mad) and the C. elegans gene Sma. SMAD proteins are signal transducers and transcriptional modulators that mediate multiple signaling pathways.
Iron-binding proteins are carrier proteins and metalloproteins that are important in iron metabolism and the immune response. Iron is required for life.

Soluble guanylyl cyclase (sGC) is the only known receptor for nitric oxide, NO. It is soluble, i.e. completely intracellular. Most notably, this enzyme is involved in vasodilation. In humans, it is encoded by the genes GUCY1A2, GUCY1A3, GUCY1B2 and GUCY1B3.

Cystathionine-β-synthase, also known as CBS, is an enzyme (EC 4.2.1.22) that in humans is encoded by the CBS gene. It catalyzes the first step of the transsulfuration pathway, from homocysteine to cystathionine:

Isopenicillin N synthase (IPNS) is a non-heme iron protein belonging to the 2-oxoglutarate (2OG)-dependent dioxygenases oxidoreductase family. This enzyme catalyzes the formation of isopenicillin N from δ-(L-α-aminoadipoyl)-L-cysteinyl-D-valine (LLD-ACV).
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.

Histidine-rich glycoprotein (HRG) is a glycoprotein that in humans is encoded by the HRG gene. The HRG protein is produced in the liver, and it could also be synthesized by monocytes, macrophages, and megakaryocytes. It possesses a multi-domain structure, which makes it capable of binding to numerous ligands and modulating various biological processes including immunity, vascularization and coagulation.

Tripartite motif-containing 24 (TRIM24) also known as transcriptional intermediary factor 1α (TIF1α) is a protein that, in humans, is encoded by the TRIM24 gene.
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine triphosphate (ATP) generation to xenobiotic degradation, the use of dioxygen as a biological oxidant is widespread and varied in the exact mechanism of its use. Enzymes employ many different schemes to use dioxygen, and this largely depends on the substrate and reaction at hand.

A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. Generally, the PAS domain acts as a molecular sensor, whereby small molecules and other proteins associate via binding of the PAS domain. Due to this sensing capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor.
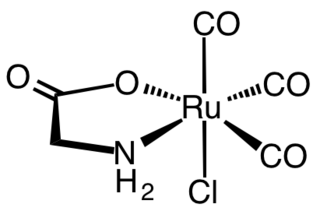
Carbon monoxide-releasing molecules (CORMs) are chemical compounds designed to release controlled amounts of carbon monoxide (CO). CORMs are being developed as potential therapeutic agents to locally deliver CO to cells and tissues, thus overcoming limitations of CO gas inhalation protocols.
A transition metal imidazole complex is a coordination complex that has one or more imidazole ligands. Complexes of imidazole itself are of little practical importance. In contrast, imidazole derivatives, especially histidine, are pervasive ligands in biology where they bind metal cofactors.

Transition metal porphyrin complexes are a family of coordination complexes of the conjugate base of porphyrins. Iron porphyrin complexes occur widely in Nature, which has stimulated extensive studies on related synthetic complexes. The metal-porphyrin interaction is a strong one such that metalloporphyrins are thermally robust. They are catalysts and exhibit rich optical properties, although these complexes remain mainly of academic interest.
Frances Ann Walker was an American chemist known for her work on heme protein chemistry. She was an elected fellow of the American Association for the Advancement of Science and the American Chemical Society.
Regulator of CO Metabolism (RcoM) is a heme-containing transcription factor found in bacteria that senses carbon monoxide (CO). In the presence of carbon monoxide, this protein upregulates expression of genes involved in carbon monoxide oxidation or carbon monoxide stress response. RcoM is functionally related to another heme-containing transcription factor, CooA, but RcoM shares no structural relationship with CooA. RcoM is composed of an N-terminal Per-Arnt-Sim (PAS) domain and a C-terminal LytTR domain. The PAS domain binds a single molecule of heme and the LytTR domain binds to DNA upstream of carbon monoxide oxidation genes. The RcoM homolog from Paraburkholderia xenovorans is known to be dimeric and binds heme using a histidine and a methionine ligand in the Fe(II) oxidation state. Carbon monoxide replaces the methionine ligand and binds directly to the heme to active RcoM for DNA binding. Relative to other heme-containing proteins, RcoM has an extraordinarily high CO affinity, with a Kd < 100 pM, allowing this protein to sense very low levels of carbon monoxide.
References
- ↑ H. Körner; H. J. Sofia; W. G. Zumft (2003). "Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs". FEMS Microbiology Reviews. 27 (5): 559–592. doi: 10.1016/S0168-6445(03)00066-4 . PMID 14638413.
- ↑ T. Shimizu; D. Huang; F. Yan; M. Stranava; M. Bartosova; V. Fojtíková; M. Martínková (2015). "Gaseous O2, NO, and CO in Signal Transduction: Structure and Function Relationships of Heme-Based Gas Sensors and Heme-Redox Sensors". Chem. Rev. 115 (13): 6491–6533. doi:10.1021/acs.chemrev.5b00018. PMID 26021768.
- ↑ Shelver, Daniel; Thorsteinsson, Marc V.; Kerby, Robert L.; Chung, Soo-Yeol; Roberts, Gary P.; Reynolds, Mark F.; Parks, Ryan B.; Burstyn, Judith N. (1999-03-01). "Identification of Two Important Heme Site Residues (Cysteine 75 and Histidine 77) in CooA, the CO-Sensing Transcription Factor of Rhodospirillum rubrum". Biochemistry. 38 (9): 2669–2678. doi:10.1021/bi982658j. ISSN 0006-2960. PMID 10052937.
- ↑ Dhawan, Ish K.; Shelver, Daniel; Thorsteinsson, Marc V.; Roberts, Gary P.; Johnson, Michael K. (1999-09-01). "Probing the Heme Axial Ligation in the CO-Sensing CooA Protein with Magnetic Circular Dichroism Spectroscopy". Biochemistry. 38 (39): 12805–12813. doi:10.1021/bi991303c. ISSN 0006-2960. PMID 10504250.
- ↑ Clark, Robert W.; Youn, Hwan; Parks, Ryan B.; Cherney, Melisa M.; Roberts, Gary P.; Burstyn, Judith N. (2004-11-01). "Investigation of the Role of the N-Terminal Proline, the Distal Heme Ligand in the CO Sensor CooA". Biochemistry. 43 (44): 14149–14160. doi:10.1021/bi0487948. ISSN 0006-2960. PMID 15518565.
- ↑ Inagaki, Sayaka; Masuda, Chiaki; Akaishi, Tetsuhiro; Nakajima, Hiroshi; Yoshioka, Shiro; Ohta, Takehiro; Pal, Biswajit; Kitagawa, Teizo; Aono, Shigetoshi (February 2005). "Spectroscopic and Redox Properties of a CooA Homologue from Carboxydothermus hydrogenoformans". Journal of Biological Chemistry. 280 (5): 3269–3274. doi: 10.1074/jbc.m409884200 . ISSN 0021-9258. PMID 15537640.
- ↑ G. P. Roberts; R. L. Kerby; H. Youn; M. Conrad (2005). "CooA, A Paradigm for Gas Sensing Regulatory Proteins". J. Inorg. Biochem. 99 (1): 280–92. doi:10.1016/j.jinorgbio.2004.10.032. PMID 15598507.
- ↑ Shigetoshi Aono (2003). "Biochemical and Biophysical Properties of the CO-Sensing Transcriptional Activator CooA". Acc. Chem. Res. 36 (11): 825–31. doi:10.1021/ar020097p. PMID 14622029.
- ↑ Lanzilotta, William N.; Schuller, David J.; Thorsteinsson, Marc V.; Kerby, Robert L.; Roberts, Gary P.; Poulos, Thomas L. (October 2000). "Structure of the CO sensing transcription activator CooA". Nature Structural Biology. 7 (10): 876–880. doi:10.1038/82820. ISSN 1545-9985. PMID 11017196. S2CID 26285016.
- ↑ Komori, Hirofumi; Inagaki, Sayaka; Yoshioka, Shiro; Aono, Shigetoshi; Higuchi, Yoshiki (2007-03-30). "Crystal Structure of CO-sensing Transcription Activator CooA Bound to Exogenous Ligand Imidazole". Journal of Molecular Biology. 367 (3): 864–871. doi:10.1016/j.jmb.2007.01.043. ISSN 0022-2836. PMID 17292914.
- ↑ Borjigin, M.; Li, H.; Lanz, N. D.; Kerby, R. L.; Roberts, G. P.; Poulos, T. L. (2007-03-01). "Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA". Acta Crystallographica Section D: Biological Crystallography. 63 (3): 282–287. Bibcode:2007AcCrD..63..282B. doi:10.1107/S0907444906051638. ISSN 0907-4449. PMID 17327664.