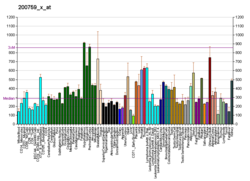
GATA-binding factor 1 or GATA-1 is the founding member of the GATA family of transcription factors. This protein is widely expressed throughout vertebrate species. In humans and mice, it is encoded by the GATA1 and Gata1 genes, respectively. These genes are located on the X chromosome in both species.
In molecular genetics, the Krüppel-like family of transcription factors (KLFs) are a set of eukaryotic C2H2 zinc finger DNA-binding proteins that regulate gene expression. This family has been expanded to also include the Sp transcription factor and related proteins, forming the Sp/KLF family.

Runt-related transcription factor 2 (RUNX2) also known as core-binding factor subunit alpha-1 (CBF-alpha-1) is a protein that in humans is encoded by the RUNX2 gene. RUNX2 is a key transcription factor associated with osteoblast differentiation.
CCAAT/enhancer-binding protein beta is a protein that in humans is encoded by the CEBPB gene.

Nuclear factor erythroid 2-related factor 2 (NRF2), also known as nuclear factor erythroid-derived 2-like 2, is a transcription factor that in humans is encoded by the NFE2L2 gene. NRF2 is a basic leucine zipper (bZIP) protein that may regulate the expression of antioxidant proteins that protect against oxidative damage triggered by injury and inflammation, according to preliminary research. In vitro, NRF2 binds to antioxidant response elements (AREs) in the promoter regions of genes encoding cytoprotective proteins. NRF2 induces the expression of heme oxygenase 1 in vitro leading to an increase in phase II enzymes. NRF2 also inhibits the NLRP3 inflammasome.

Homeobox protein Hox-A9 is a protein that in humans is encoded by the HOXA9 gene.

Nuclear respiratory factor 1, also known as Nrf1, Nrf-1, NRF1 and NRF-1, encodes a protein that homodimerizes and functions as a transcription factor which activates the expression of some key metabolic genes regulating cellular growth and nuclear genes required for respiration, heme biosynthesis, and mitochondrial DNA transcription and replication. The protein has also been associated with the regulation of neurite outgrowth. Alternate transcriptional splice variants, which encode the same protein, have been characterized. Additional variants encoding different protein isoforms have been described but they have not been fully characterized. Confusion has occurred in bibliographic databases due to the shared symbol of NRF1 for this gene and for "nuclear factor -like 1" which has an official symbol of NFE2L1.

Interleukin enhancer-binding factor 3 is a protein that in humans is encoded by the ILF3 gene.

X-box binding protein 1, also known as XBP1, is a protein which in humans is encoded by the XBP1 gene. The XBP1 gene is located on chromosome 22 while a closely related pseudogene has been identified and localized to chromosome 5. The XBP1 protein is a transcription factor that regulates the expression of genes important to the proper functioning of the immune system and in the cellular stress response.

DNA damage-inducible transcript 3, also known as C/EBP homologous protein (CHOP), is a pro-apoptotic transcription factor that is encoded by the DDIT3 gene. It is a member of the CCAAT/enhancer-binding protein (C/EBP) family of DNA-binding transcription factors. The protein functions as a dominant-negative inhibitor by forming heterodimers with other C/EBP members, preventing their DNA binding activity. The protein is implicated in adipogenesis and erythropoiesis and has an important role in the cell's stress response.
cAMP responsive element modulator is a protein that in humans is encoded by the CREM gene, and it belongs to the cAMP-responsive element binding protein family. It has multiple isoforms, which act either as repressors or activators. CREB family is important for in regulating transcription in response to various stresses, metabolic and developmental signals. CREM transcription factors also play an important role in many physiological systems, such as cardiac function, circadian rhythms, locomotion and spermatogenesis.

Transcription regulator protein BACH1 is a protein that in humans is encoded by the BACH1 gene.

Transcription factor NF-E2 45 kDa subunit is a protein that in humans is encoded by the NFE2 gene.
Transcription factor MafG is a bZip Maf transcription factor protein that in humans is encoded by the MAFG gene.

Transcription factor MafK is a bZip Maf transcription factor protein that in humans is encoded by the MAFK gene.

Basic leucine zipper transcription factor, ATF-like, also known as BATF, is a protein which in humans is encoded by the BATF gene.

Transcription factor MafF is a bZip Maf transcription factor protein that in humans is encoded by the MAFF gene.

Nuclear factor, interleukin 3 regulated, also known as NFIL3 or E4BP4 is a protein which in humans is encoded by the NFIL3 gene.

Nuclear factor -like factor 3, also known as NFE2L3 or 'NRF3', is a transcription factor that in humans is encoded by the Nfe2l3 gene.
Small Maf proteins are basic region leucine zipper-type transcription factors that can bind to DNA and regulate gene regulation. There are three small Maf (sMaf) proteins, namely MafF, MafG, and MafK, in vertebrates. HUGO Gene Nomenclature Committee (HGNC)-approved gene names of MAFF, MAFG and MAFK are “v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F, G, and K”, respectively.