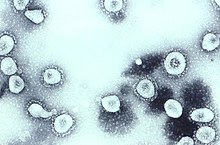
Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.

Severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), previously known as severe acute respiratory syndrome coronavirus (SARS-CoV), is a strain of coronavirus that causes severe acute respiratory syndrome (SARS), the respiratory illness responsible for the 2002–2004 SARS outbreak. It is an enveloped, positive-sense, single-stranded RNA virus that infects the epithelial cells within the lungs. The virus enters the host cell by binding to angiotensin-converting enzyme 2. It infects humans, bats, and palm civets. The SARS-CoV-1 outbreak was largely brought under control by simple public health measures. Testing people with symptoms, isolating and quarantining suspected cases, and restricting travel all had an effect. SARS-CoV-1 was most transmissible when patients were sick, so its spread could be effectively suppressed by isolating patients with symptoms.

Human coronavirus NL63 (HCoV-NL63) is a species of coronavirus, specifically a Setracovirus from among the Alphacoronavirus genus. It was identified in late 2004 in patients in the Netherlands by Lia van der Hoek and Krzysztof Pyrc using a novel virus discovery method VIDISCA. Later on the discovery was confirmed by the researchers from the Rotterdam, the Netherlands The virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to ACE2. Infection with the virus has been confirmed worldwide, and has an association with many common symptoms and diseases. Associated diseases include mild to moderate upper respiratory tract infections, severe lower respiratory tract infection, croup and bronchiolitis.

Murine coronavirus (M-CoV) is a virus in the genus Betacoronavirus that infects mice. Belonging to the subgenus Embecovirus, murine coronavirus strains are enterotropic or polytropic. Enterotropic strains include mouse hepatitis virus (MHV) strains D, Y, RI, and DVIM, whereas polytropic strains, such as JHM and A59, primarily cause hepatitis, enteritis, and encephalitis. Murine coronavirus is an important pathogen in the laboratory mouse and the laboratory rat. It is the most studied coronavirus in animals other than humans, and has been used as an animal disease model for many virological and clinical studies.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.
Bovine coronavirus is a coronavirus which is a member of the species Betacoronavirus 1. The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the N-acetyl-9-O-acetylneuraminic acid recepter. Infection causes calf enteritis and contributes to the enzootic pneumonia complex in calves. It can also cause winter dysentery in adult cattle. It can infect both domestic and wild ruminants and has a worldwide distribution. Transmission is horizontal, via oro-fecal or respiratory routes. Like other coronaviruses from genus Betacoronavirus, subgenus Embecovirus, it has a surface protein called hemagglutinin esterase (HE) in addition to the four structural proteins shared by all coronaviruses.

Middle East respiratory syndrome–related coronavirus (MERS-CoV), or EMC/2012 (HCoV-EMC/2012), is the virus that causes Middle East respiratory syndrome (MERS). It is a species of coronavirus which infects humans, bats, and camels. The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the DPP4 receptor. The species is a member of the genus Betacoronavirus and subgenus Merbecovirus.

The 1889–1890 pandemic, often referred to as the "Asiatic flu" or "Russian flu", was a worldwide respiratory viral pandemic. It was the last great pandemic of the 19th century, and is among the deadliest pandemics in history. The pandemic killed about 1 million people out of a world population of about 1.5 billion. The most reported effects of the pandemic took place from October 1889 to December 1890, with recurrences in March to June 1891, November 1891 to June 1892, the northern winter of 1893–1894, and early 1895.
Novel coronavirus (nCoV) is a provisional name given to coronaviruses of medical significance before a permanent name is decided upon. Although coronaviruses are endemic in humans and infections normally mild, such as the common cold, cross-species transmission has produced some unusually virulent strains which can cause viral pneumonia and in serious cases even acute respiratory distress syndrome and death.

Human coronavirus HKU1 (HCoV-HKU1) is a species of coronavirus in humans and animals. It causes an upper respiratory disease with symptoms of the common cold, but can advance to pneumonia and bronchiolitis. It was first discovered in January 2004 from one man in Hong Kong. Subsequent research revealed it has global distribution and earlier genesis.

Betacoronavirus is one of four genera of coronaviruses. Member viruses are enveloped, positive-strand RNA viruses that infect mammals, including humans. The natural reservoir for betacoronaviruses are bats and rodents. Rodents are the reservoir for the subgenus Embecovirus, while bats are the reservoir for the other subgenera.

Betacoronavirus 1 is a species of coronavirus which infects humans and cattle. The infecting virus is an enveloped, positive-sense, single-stranded RNA virus and is a member of the genus Betacoronavirus and subgenus Embecovirus. Like other embecoviruses, it has an additional shorter spike-like surface protein called hemagglutinin esterase (HE) as well as the larger coronavirus spike protein.

Human coronavirus 229E (HCoV-229E) is a species of coronavirus which infects humans and bats. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor. Along with Human coronavirus OC43, it is one of the viruses responsible for the common cold. HCoV-229E is a member of the genus Alphacoronavirus and subgenus Duvinacovirus.
Tylonycteris bat coronavirus HKU4 is an enveloped, positive-sense single-stranded RNA virus mammalian Group 2 Betacoronavirus that has been found to be genetically related to the Middle East respiratory syndrome-related coronavirus (MERS-CoV) that is responsible for the 2012 Middle East respiratory syndrome coronavirus outbreak in Saudi Arabia, Jordan, United Arab Emirates, the United Kingdom, France, and Italy.
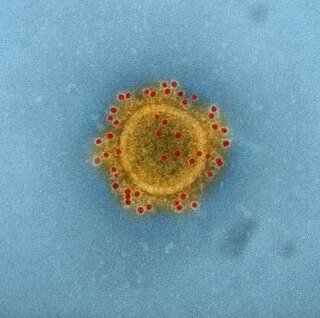
MERS coronavirus EMC/2012 is a strain of coronavirus isolated from the sputum of the first person to become infected with what was later named Middle East respiratory syndrome–related coronavirus (MERS-CoV), a virus that causes Middle East respiratory syndrome (MERS).

Embecovirus is a subgenus of coronaviruses in the genus Betacoronavirus. The viruses in this subgenus, unlike other coronaviruses, have a hemagglutinin esterase (HE) gene. The viruses in the subgenus were previously known as group 2a coronaviruses.

Coronavirus diseases are caused by viruses in the coronavirus subfamily, a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, the group of viruses cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19. As of 2021, 45 species are registered as coronaviruses, whilst 11 diseases have been identified, as listed below.

The history of coronaviruses is an account of the discovery of the diseases caused by coronaviruses and the diseases they cause. It starts with the first report of a new type of upper-respiratory tract disease among chickens in North Dakota, U.S., in 1931. The causative agent was identified as a virus in 1933. By 1936, the disease and the virus were recognised as unique from other viral disease. They became known as infectious bronchitis virus (IBV), but later officially renamed as Avian coronavirus.

ORF3a is a gene found in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It encodes an accessory protein about 275 amino acid residues long, which is thought to function as a viroporin. It is the largest accessory protein and was the first of the SARS-CoV accessory proteins to be described.