Isomerases are a general class of enzymes that convert a molecule from one isomer to another. Isomerases facilitate intramolecular rearrangements in which bonds are broken and formed. The general form of such a reaction is as follows:
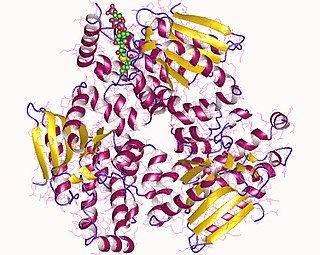
Enoyl-CoA-(∆) isomerase (EC 5.3.3.8, also known as dodecenoyl-CoA- isomerase, 3,2-trans-enoyl-CoA isomerase, ∆3 ,∆2 -enoyl-CoA isomerase, or acetylene-allene isomerase, is an enzyme that catalyzes the conversion of cis- or trans-double bonds of coenzyme A bound fatty acids at gamma-carbon to trans double bonds at beta-carbon as below:
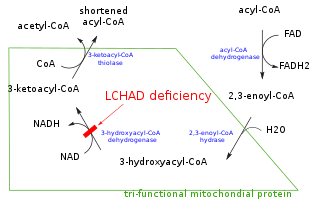
In biochemistry and metabolism, beta oxidation (also β-oxidation) is the catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA, which enters the citric acid cycle, and NADH and FADH2, which are co-enzymes used in the electron transport chain. It is named as such because the beta carbon of the fatty acid undergoes oxidation to a carbonyl group. Beta-oxidation is primarily facilitated by the mitochondrial trifunctional protein, an enzyme complex associated with the inner mitochondrial membrane, although very long chain fatty acids are oxidized in peroxisomes.

Methylmalonyl-CoA mutase is a mitochondrial homodimer apoenzyme that focuses on the catalysis of methylmalonyl CoA to succinyl CoA. The enzyme is bound to adenosylcobalamin, a hormonal derivative of vitamin B12 in order to function. Methylmalonyl-CoA mutase deficiency is caused by genetic defect in the MUT gene responsible for encoding the enzyme. Deficiency in this enzyme accounts for 60% of the cases of methylmalonic acidemia.

Phosphoglycerate mutase (PGM) is any enzyme that catalyzes step 8 of glycolysis - the internal transfer of a phosphate group from C-3 to C-2 which results in the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG) through a 2,3-bisphosphoglycerate intermediate. These enzymes are categorized into the two distinct classes of either cofactor-dependent (dPGM) or cofactor-independent (iPGM). The dPGM enzyme is composed of approximately 250 amino acids and is found in all vertebrates as well as in some invertebrates, fungi, and bacteria. The iPGM class is found in all plants and algae as well as in some invertebrate, fungi, and Gram-positive bacteria. This class of PGM enzyme shares the same superfamily as alkaline phosphatase.

Methylmalonyl-CoA mutase (EC 5.4.99.2, MCM), mitochondrial, also known as methylmalonyl-CoA isomerase, is a protein that in humans is encoded by the MUT gene. This vitamin B12-dependent enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA in humans. Mutations in MUT gene may lead to various types of methylmalonic aciduria.

In enzymology, a nicotinate dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, a beta-lysine 5,6-aminomutase is an enzyme that catalyzes the chemical reaction

In enzymology, D-lysine 5,6-aminomutase is an enzyme that catalyzes the chemical reaction
In enzymology, a D-ornithine 4,5-aminomutase is an enzyme that catalyzes the chemical reaction
In enzymology, a leucine 2,3-aminomutase is an enzyme that catalyzes the chemical reaction
In enzymology, a methylaspartate mutase is an enzyme that catalyzes the chemical reaction
In enzymology, a methylitaconate Δ-isomerase is an enzyme that catalyzes the chemical reaction

In enzymology, a phosphoacetylglucosamine mutase is an enzyme that catalyzes the chemical reaction
In enzymology, a phosphoglucosamine mutase is an enzyme that catalyzes the chemical reaction
In enzymology, a tyrosine 2,3-aminomutase is an enzyme that catalyzes the chemical reaction
The enzyme ethanolamine ammonia-lyase (EC 4.3.1.7) catalyzes the chemical reaction

Glutathione S-transferase Zeta 1 is an enzyme that in humans is encoded by the GSTZ1 gene on chromosome 14.
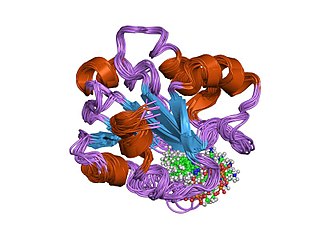
In molecular biology, the vitamin B12-binding domain is a protein domain which binds to cobalamin. It can bind two different forms of the cobalamin cofactor, with cobalt bonded either to a methyl group (methylcobalamin) or to 5'-deoxyadenosine (adenosylcobalamin). Cobalamin-binding domains are mainly found in two families of enzymes present in animals and prokaryotes, which perform distinct kinds of reactions at the cobalt-carbon bond. Enzymes that require methylcobalamin carry out methyl transfer reactions. Enzymes that require adenosylcobalamin catalyse reactions in which the first step is the cleavage of adenosylcobalamin to form cob(II)alamin and the 5'-deoxyadenosyl radical, and thus act as radical generators. In both types of enzymes the B12-binding domain uses a histidine to bind the cobalt atom of cobalamin cofactors. This histidine is embedded in a DXHXXG sequence, the most conserved primary sequence motif of the domain. Proteins containing the cobalamin-binding domain include:

In enzymology, a xylose isomerase is an enzyme that catalyzes the interconversion of D-xylose and D-xylulose. This enzyme belongs to the family of isomerases, specifically those intramolecular oxidoreductases interconverting aldoses and ketoses. The isomerase has now been observed in nearly a hundred species of bacteria. Xylose-isomerases are also commonly called fructose-isomerases due to their ability to interconvert glucose and fructose. The systematic name of this enzyme class is D-xylose aldose-ketose-isomerase. Other names in common use include D-xylose isomerase, D-xylose ketoisomerase, and D-xylose ketol-isomerase.










