
Luciferase is a generic term for the class of oxidative enzymes that produce bioluminescence, and is usually distinguished from a photoprotein. The name was first used by Raphaël Dubois who invented the words luciferin and luciferase, for the substrate and enzyme, respectively. Both words are derived from the Latin word lucifer, meaning "lightbearer", which in turn is derived from the Latin words for "light" (lux) and "to bring or carry" (ferre).
An oxygenase is any enzyme that oxidizes a substrate by transferring the oxygen from molecular oxygen O2 (as in air) to it. The oxygenases form a class of oxidoreductases; their EC number is EC 1.13 or EC 1.14.

Firefly luciferase is the light-emitting enzyme responsible for the bioluminescence of fireflies and click beetles. The enzyme catalyses the oxidation of firefly luciferin, requiring oxygen and ATP. Because of the requirement of ATP, firefly luciferases have been used extensively in biotechnology.
In enzymology, a 4-aminobenzoate 1-monooxygenase (EC 1.14.13.27) is an enzyme that catalyzes the chemical reaction
In enzymology, an alkanal monooxygenase (FMN-linked) (EC 1.14.14.3) is an enzyme that catalyzes the chemical reaction

In enzymology, a camphor 5-monooxygenase (EC 1.14.15.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a choline monooxygenase (EC 1.14.15.7) is an enzyme that catalyzes the chemical reaction
In enzymology, a Latia-luciferin monooxygenase (demethylating) (EC 1.14.99.21) is an enzyme that catalyzes the chemical reaction
In enzymology, a salicylate 1-monooxygenase (EC 1.14.13.1) is an enzyme that catalyzes the chemical reaction
Arginine 2-monooxygenase (EC 1.13.12.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a lactate 2-monooxygenase (EC 1.13.12.4) is an enzyme that catalyzes the chemical reaction
In enzymology, a lysine 2-monooxygenase (EC 1.13.12.2) is an enzyme that catalyzes the chemical reaction
In enzymology, an Oplophorus-luciferin 2-monooxygenase, also known as Oplophorus luciferase is a luciferase, an enzyme, from the deep-sea shrimp Oplophorus gracilirostris [2], belonging to a group of coelenterazine luciferases. Unlike other luciferases, it has a broader substrate specificity [3,4,6] and can also bind to bisdeoxycoelenterazine efficiently [3,4]. It is the third example of a luciferase to be purified in lab [2]. The systematic name of this enzyme class is Oplophorus-luciferin:oxygen 2-oxidoreductase (decarboxylating). This enzyme is also called Oplophorus luciferase.
In enzymology, a phenylalanine 2-monooxygenase (EC 1.13.12.9) is an enzyme that catalyzes the chemical reaction
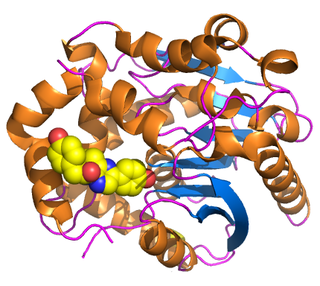
Renilla-luciferin 2-monooxygenase, Renilla luciferase, or RLuc, is a bioluminescent enzyme found in Renilla reniformis, belonging to a group of coelenterazine luciferases. Of this group of enzymes, the luciferase from Renilla reniformis has been the most extensively studied, and due to its bioluminescence requiring only molecular oxygen, has a wide range of applications, with uses as a reporter gene probe in cell culture, in vivo imaging, and various other areas of biological research. Recently, chimeras of RLuc have been developed and demonstrated to be the brightest luminescent proteins to date, and have proved effective in both noninvasive single-cell and whole body imaging.
In enzymology, a tryptophan 2-monooxygenase (EC 1.13.12.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a Watasenia-luciferin 2-monooxygenase (EC 1.13.12.8) is an enzyme that catalyzes the chemical reaction

Vargulin, also called Cypridinid luciferin, Cypridina luciferin, or Vargula luciferin, is the luciferin found in the ostracod Cypridina hilgendorfii, also named Vargula hilgendorfii. These bottom dwelling ostracods emit a light stream into water when disturbed presumably to deter predation. Vargulin is also used by the midshipman fish, Porichthys.

Vargula hilgendorfii, sometimes called the sea-firefly and one of three bioluminescent species known in Japan as umi-hotaru (海蛍), is a species of ostracod crustacean. It is the only member of genus Vargula to inhabit Japanese waters; all other members of its genus inhabit the Gulf of Mexico, the Caribbean Sea, and waters off the coast of California. V. hilgendorfii was formerly more common, but its numbers have fallen significantly.
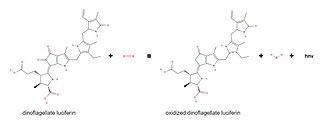
Dinoflagellate luciferase (EC 1.13.12.18, Gonyaulax luciferase) is a specific luciferase, an enzyme with systematic name dinoflagellate-luciferin:oxygen 132-oxidoreductase.







