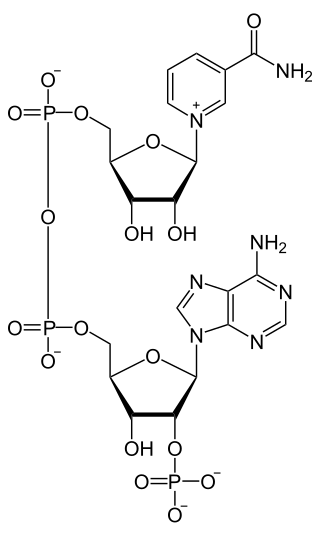
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form of NADP+, the oxidized form. NADP+ is used by all forms of cellular life.
Thioredoxin reductases are enzymes that reduce thioredoxin (Trx). Two classes of thioredoxin reductase have been identified: one class in bacteria and some eukaryotes and one in animals. In bacteria TrxR also catalyzes the reduction of glutaredoxin like proteins known as NrdH. Both classes are flavoproteins which function as homodimers. Each monomer contains a FAD prosthetic group, a NADPH binding domain, and an active site containing a redox-active disulfide bond.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.
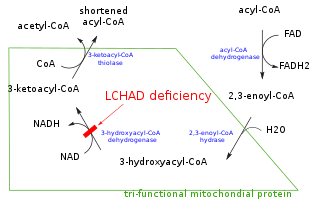
In biochemistry and metabolism, beta oxidation (also β-oxidation) is the catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA, which enters the citric acid cycle, and NADH and FADH2, which are co-enzymes used in the electron transport chain. It is named as such because the beta carbon of the fatty acid undergoes oxidation to a carbonyl group. Beta-oxidation is primarily facilitated by the mitochondrial trifunctional protein, an enzyme complex associated with the inner mitochondrial membrane, although very long chain fatty acids are oxidized in peroxisomes.
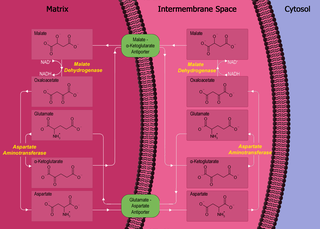
The malate-aspartate shuttle is a biochemical system for translocating electrons produced during glycolysis across the semipermeable inner membrane of the mitochondrion for oxidative phosphorylation in eukaryotes. These electrons enter the electron transport chain of the mitochondria via reduction equivalents to generate ATP. The shuttle system is required because the mitochondrial inner membrane is impermeable to NADH, the primary reducing equivalent of the electron transport chain. To circumvent this, malate carries the reducing equivalents across the membrane.

Succinate dehydrogenase complex, subunit A, flavoprotein variant is a protein that in humans is encoded by the SDHA gene. This gene encodes a major catalytic subunit of succinate-ubiquinone oxidoreductase, a complex of the mitochondrial respiratory chain. The complex is composed of four nuclear-encoded subunits and is localized in the mitochondrial inner membrane. SDHA contains the FAD binding site where succinate is deprotonated and converted to fumarate. Mutations in this gene have been associated with a form of mitochondrial respiratory chain deficiency known as Leigh Syndrome. A pseudogene has been identified on chromosome 3q29. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.

The glycerol-3-phosphate shuttle is a mechanism used in skeletal muscle and the brain that regenerates NAD+ from NADH, a by-product of glycolysis. The NADH generated during glycolysis is found in the cytoplasm and must be transported into the mitochondria to enter the oxidative phosphorylation pathway. However, the inner mitochondrial membrane is impermeable to NADH and NAD+ and does not contain a transport system for these electron carriers. Either the glycerol-3-phosphate shuttle pathway or the malate-aspartate shuttle pathway, depending on the tissue of the organism, must be taken to transport cytoplasmic NADH into the mitochondria. The shuttle consists of the sequential activity of two proteins; Cytoplasmic glycerol-3-phosphate dehydrogenase (cGPD) transfers an electron pair from NADH to dihydroxyacetone phosphate (DHAP), forming glycerol-3-phosphate (G3P) and regenerating NAD+ needed to generate energy via glycolysis. The other protein, mitochondrial glycerol-3-phosphate dehydrogenase (mGPD) catalyzes the oxidation of G3P by FAD, regenerating DHAP in the cytosol and forming FADH2 in the mitochondrial matrix. In mammals, its activity in transporting reducing equivalents across the mitochondrial membrane is considered secondary to the malate-aspartate shuttle.

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction
In enzymology, an anthraniloyl-CoA monooxygenase (EC 1.14.13.40) is an enzyme that catalyzes the chemical reaction

In enzymology, a NADH peroxidase (EC 1.11.1.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction

NAD(P)H dehydrogenase [quinone] 1 is an enzyme that in humans is encoded by the NQO1 gene. This protein-coding gene is a member of the NAD(P)H dehydrogenase (quinone) family and encodes a 2-electron reductase (enzyme). This FAD-binding protein forms homodimers and performs two-electron reduction of quinones to hydroquinones and of other redox dyes. It has a preference for short-chain acceptor quinones, such as ubiquinone, benzoquinone, juglone and duroquinone. This gene has an important paralog NQO2. This protein is located in the cytosol.
NAD(P)H dehydrogenase, quinone 2, also known as QR2, is a protein that in humans is encoded by the NQO2 gene. It is a phase II detoxification enzyme which can carry out two or four electron reductions of quinones. Its mechanism of reduction is through a ping-pong mechanism involving its FAD cofactor. Initially in a reductive phase NQO2 binds to reduced dihydronicotinamide riboside (NRH) electron donor, and mediates a hydride transfer from NRH to FAD. Then, in an oxidative phase, NQO2 binds to its quinone substrate and reduces the quinone to a dihydroquinone. Besides the two catalytic FAD, NQO2 also has two zinc ions. It is not clear whether the metal has a catalytic role. NQO2 is a paralog of NQO1
NQO2 is a homodimer. NQO2 can be inhibited by resveratrol. One of QR2's binding sites responds to 2-iodomelatonin, and has been referred to as MT3.

Renalase, FAD-dependent amine oxidase is an enzyme that in humans is encoded by the RNLS gene. Renalase is a flavin adenine dinucleotide-dependent amine oxidase that is secreted into the blood from the kidney.

Diacetyl reductase ((S)-acetoin forming) (EC 1.1.1.304, (S)-acetoin dehydrogenase) is an enzyme with systematic name (S)-acetoin:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
FAD reductase (NADH) (EC 1.5.1.37, NADH-FAD reductase, NADH-dependent FAD reductase) is an enzyme with systematic name FADH2:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
6-hydroxy-3-succinoylpyridine 3-monooxygenase (EC 1.14.13.163, 6-hydroxy-3-succinoylpyridine hydroxylase, hspA (gene), hspB (gene)) is an enzyme with systematic name 4-(6-hydroxypyridin-3-yl)-4-oxobutanoate,NADH:oxygen oxidoreductase (3-hydroxylating, succinate semialdehyde releasing). This enzyme catalyses the following chemical reaction
Tryptophan 7-halogenase (EC 1.14.19.9, PrnA, RebH) is an enzyme with systematic name L-tryptophan:FADH2 oxidoreductase (7-halogenating). This enzyme catalyses the following chemical reaction:
Anthranilate 3-monooxygenase (FAD) (EC 1.14.14.8, anthranilate 3-hydroxylase, anthranilate hydroxylase) is an enzyme with systematic name anthranilate,FAD:oxygen oxidoreductase (3-hydroxylating). This enzyme catalyses the following chemical reaction
Cytochrome P450 aromatic O-demethylase is a bacterial enzyme that catalyzes the demethylation of lignin and various lignols. The net reaction follows the following stoichiometry, illustrated with a generic methoxy arene:















