
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: Duplodnaviria and Varidnaviria, and ssDNA viruses are almost exclusively assigned to the realm Monodnaviria, which also includes some dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Reverse transcribing viruses, which have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase, are classified into the kingdom Pararnavirae in the realm Riboviria.

Parvoviruses are a family of animal viruses that constitute the family Parvoviridae. They have linear, single-stranded DNA (ssDNA) genomes that typically contain two genes encoding for a replication initiator protein, called NS1, and the protein the viral capsid is made of. The coding portion of the genome is flanked by telomeres at each end that form into hairpin loops that are important during replication. Parvovirus virions are small compared to most viruses, at 23–28 nanometers in diameter, and contain the genome enclosed in an icosahedral capsid that has a rugged surface.

Vicia faba, commonly known as the broad bean, fava bean, or faba bean, is a species of vetch, a flowering plant in the pea and bean family Fabaceae. It is widely cultivated as a crop for human consumption, and also as a cover crop. Varieties with smaller, harder seeds that are fed to horses or other animals are called field bean, tic bean or tick bean. Horse bean, Vicia faba var. equinaPers., is a variety recognized as an accepted name. This legume is very common in Southern European, Northern European, East Asian, Latin American and North African cuisines.

Plant viruses are viruses that affect plants. Like all other viruses, plant viruses are obligate intracellular parasites that do not have the molecular machinery to replicate without a host. Plant viruses can be pathogenic to higher plants.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

Nanoviridae is a family of viruses. Plants serve as natural hosts. There are currently 12 species in this family, divided among 2 genera and one unassigned species. Diseases associated with this family include: stunting. Their name is derived from the Greek word νᾶνος, because of their small genome and their stunting effect on infected plants.
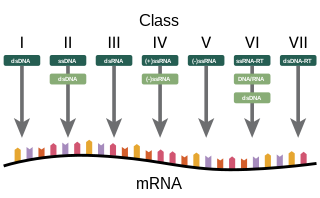
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.

Maize streak virus (MSV) is a virus primarily known for causing maize streak disease (MSD) in its major host, and which also infects over 80 wild and domesticated grasses. It is an insect-transmitted maize pathogen in the genus Mastrevirus of the family Geminiviridae that is endemic in sub-Saharan Africa and neighbouring Indian Ocean island territories such as Madagascar, Mauritius and La Reunion. The A-strain of MSV (MSV-A) causes sporadic maize streak disease epidemics throughout the maize growing regions of Africa. MSV was first described by the South African entomologist Claude Fuller who referred to it in a 1901 report as "mealie variegation".
Asparagus virus 1 (AV-1) is one of the nine known viruses that affects asparagus plants. It is in the Potyviridae family. Initially reported by G. L Hein in 1960, it is a member of the genus Potyvirus and causes no distinct symptoms in asparagus plants. The only known plant that can get AV-1 is asparagus plants. It is spread by aphids vectors, which means that aphids do not cause the AV-1, but they do spread it.

Tobacco streak virus (TSV) is a plant pathogenic virus of the family Bromoviridae, in the genus Ilarvirus. It has a wide host range, with at least 200 susceptible species. TSV is generally more problematic in the tropics or warmer climates. TSV does not generally lead to epidemics, with the exception of sunflowers in India and Australia, and peanuts in India.

The black bean aphid is a small black insect in the genus Aphis, with a broad, soft body, a member of the order Hemiptera. Other common names include blackfly, bean aphid, and beet leaf aphid. In the warmer months of the year, it is found in large numbers on the undersides of leaves and on the growing tips of host plants, including various agricultural crops and many wild and ornamental plants. Both winged and wingless forms exist, and at this time of year, they are all females. They suck sap from stems and leaves and cause distortion of the shoots, stunted plants, reduced yield, and spoiled crops. This aphid also acts as a vector for viruses that cause plant disease, and the honeydew it secretes may encourage the growth of sooty mould. It breeds profusely by live birth, but its numbers are kept in check, especially in the later part of the summer, by various predatory and parasitic insects. Ants feed on the honeydew it produces, and take active steps to remove the aphid's enemies. It is a widely distributed pest of agricultural crops and can be controlled by chemical or biological means. In the autumn, winged forms move to different host plants, where both males and females are produced. These mate and the females lay eggs which overwinter.
Alphasatellites are a single-stranded DNA family of satellite viruses that depend on the presence of another virus to replicate their genomes. As such, they have minimal genomes with very low genomic redundancy. The genome is a single circular single strand DNA molecule. The first alphasatellites were described in 1999 and were associated with cotton leaf curl disease and Ageratum yellow vein disease. As begomoviruses are being characterised at the molecular level an increasing number of alphasatellites are being described.

Nanovirus is a genus of viruses, in the family Nanoviridae. Legume plants serve as natural hosts. There are 11 species in this genus. Diseases associated with this genus include: stunting, severe necrosis and early plant death.

Genomoviridae is a family of single stranded DNA viruses that mainly infect fungi. The genomes of this family are small. The genomes are circular single-stranded DNA and encode rolling-circle replication initiation proteins (Rep) and unique capsid proteins. In Rep-based phylogenies, genomoviruses form a sister clade to plant viruses of the family Geminiviridae. Ten genera are recognized in this family.
In virology, realm is the highest taxonomic rank established for viruses by the International Committee on Taxonomy of Viruses (ICTV), which oversees virus taxonomy. Six virus realms are recognized and united by specific highly conserved traits:

HUH endonucleases (HUH-tags) are sequence-specific single-stranded DNA (ssDNA) binding proteins originating from numerous species of bacteria and viruses. Viral HUH endonucleases are involved in initiating rolling circle replication while ones of bacterial origin initiate bacterial conjugation. In biotechnology, they can be used to create protein-DNA linkages, akin to other methods such as SNAP-tag. In doing so, they create a 5' covalent bond between the ssDNA and the protein. HUH endonucleases can be fused with other proteins or used as protein tags.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.

Cressdnaviricota is a phylum of viruses with small, circular single-stranded DNA genomes and encoding rolling circle replication-initiation proteins with the N-terminal HUH endonuclease and C-terminal superfamily 3 helicase domains.

Bacilladnaviridae is a family of single-stranded DNA viruses that primarily infect diatoms.
Rolling hairpin replication (RHR) is a unidirectional, strand displacement form of DNA replication used by parvoviruses, a group of viruses that constitute the family Parvoviridae. Parvoviruses have linear, single-stranded DNA (ssDNA) genomes in which the coding portion of the genome is flanked by telomeres at each end that form hairpin loops. During RHR, these hairpin loops repeatedly unfold and refold to change the direction of DNA replication so that replication progresses in a continuous manner back and forth across the genome. RHR is initiated and terminated by an endonuclease encoded by parvoviruses that is variously called NS1 or Rep, and RHR is similar to rolling circle replication, which is used by ssDNA viruses that have circular genomes.












