
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are 1500-1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.

Corynebacterium diphtheriae is the pathogenic bacterium that causes diphtheria. It is also known as the Klebs–Löffler bacillus, because it was discovered in 1884 by German bacteriologists Edwin Klebs (1834–1912) and Friedrich Löffler (1852–1915). The bacteria are usually harmless unless they are infected by a bacteriophage that carries a gene that gives rise to a toxin. This toxin causes the disease. Diphtheria is caused by the adhesion and infiltration of the bacteria into the mucosal layers of the body, primarily affecting the respiratory tract and the subsequent release of an endotoxin. The toxin has a localized effect on skin lesions, as well as a metastatic, proteolytic effects on other organ systems in severe infections. Originally a major cause of childhood mortality, diphtheria has been almost entirely eradicated due to the vigorous administration of the diphtheria vaccination in the 1910s.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

Tet Repressor proteins are proteins playing an important role in conferring antibiotic resistance to large categories of bacterial species.
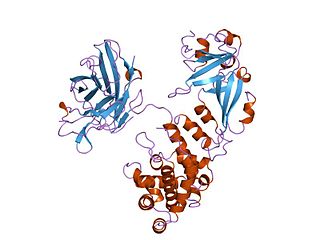
Diphtheria toxin is an exotoxin secreted by mainly by Corynebacterium diphtheriae but also by Corynebacterium ulcerans and Corynebacterium pseudotuberculosis. the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
In the field of molecular biology the 6S RNA is a non-coding RNA that was one of the first to be identified and sequenced. What it does in the bacterial cell was unknown until recently. In the early 2000s scientists found out the function of 6S RNA to be as a regulator of sigma 70-dependent gene transcription. All bacterial RNA polymerases have a subunit called a sigma factor. The sigma factors are important because they control how DNA promoter binding and RNA transcription start sites. Sigma 70 was the first one to be discovered in Escherichia coli.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.
POU is a family of eukaryotic transcription factors that have well-conserved homeodomains. The Pou domain is a bipartite DNA binding domain found in these proteins.
HutP is one of the anti-terminator proteins of Bacillus subtilis, which is responsible for regulating the expression of the hut structural genes of this organism in response to changes in the intracellular levels of L-histidine and divalent metal ions. In the hut operon, HutP is located just downstream from the promoter, while the five other subsequent structural genes, hutH, hutU, hutI, hutG and hutM, are positioned far downstream from the promoter. In the presence of L-histidine and divalent metal ions, HutP binds to the nascent hut mRNA leader transcript. This allows the anti-terminator to form, thereby preventing the formation of the terminator and permitting transcriptional read-through into the hut structural genes. In the absence of L-histidine and divalent metal ions or both, HutP does not bind to the hut mRNA, thus allowing the formation of a stem loop terminator structure within the nucleotide sequence located between the hutP and structural genes.

MNT is a Max-binding protein that is encoded by the MNT gene
A corynebacteriophage is a DNA-containing bacteriophage specific for bacteria of genus Corynebacterium as its host.
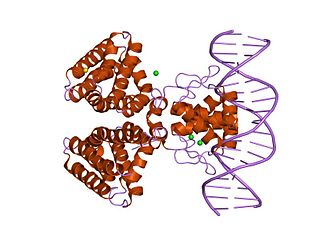
In molecular biology, the fatty acid metabolism regulator protein FadR, is a bacterial transcription factor.

In molecular biology, the ferric uptake regulator family is a family of bacterial proteins involved in regulating metal ion uptake and in metal homeostasis. The family is named for its founding member, known as the ferric uptake regulator or ferric uptake regulatory protein (Fur). Fur proteins are responsible for controlling the intracellular concentration of iron in many bacteria. Iron is essential for most organisms, but its concentration must be carefully managed over a wide range of environmental conditions; high concentrations can be toxic due to the formation of reactive oxygen species.

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.

In molecular biology, the GntR-like bacterial transcription factors are a family of transcription factors.
The nik operon is an operon required for uptake of nickel ions into the cell. It is present in many bacteria, but has been extensively studied in Helicobacter pylori. Nickel is an essential nutrient for many microorganisms, where it participates in a variety of cellular processes. However, excessive levels of nickel ions in cell can be fatal to the cell. Nickel ion concentration in the cell is regulated through the nik operon.

The Phosphate (Pho) regulon is a regulatory mechanism used for the conservation and management of inorganic phosphate within the cell. It was first discovered in Escherichia coli as an operating system for the bacterial strain, and was later identified in other species. The Pho system is composed of various components including extracellular enzymes and transporters that are capable of phosphate assimilation in addition to extracting inorganic phosphate from organic sources. This is an essential process since phosphate plays an important role in cellular membranes, genetic expression, and metabolism within the cell. Under low nutrient availability, the Pho regulon helps the cell survive and thrive despite a depletion of phosphate within the environment. When this occurs, phosphate starvation-inducible (psi) genes activate other proteins that aid in the transport of inorganic phosphate.

Protein Arginine Phosphatase (PAPs), also known as Phosphoarginine Phosphatase, is an enzyme that catalyzes the dephosphorylation of phosphoarginine residues in proteins. Protein phosphatases (PPs) are "obligatory heteromers" made up of two maximum catalytic subunits attached to a non-catalytic subunit. Arginine modification is a post-translational protein modification in gram-positive bacteria. McsB and YwIE were recently identified as phosphorylating enzymes in Bacillus Subtilis (B.Subtilis). YwIE was thought to be a protein-tyrosine-phosphatase, and McsB a tyrosine-kinase, however in 2012 Elsholz et al. showed that McsB is a protein-arginine-kinase (PAK) and YwlE is a phosphatase-arginine-phosphatase (PAP).















