
The Marchantiophyta are a division of non-vascular land plants commonly referred to as hepatics or liverworts. Like mosses and hornworts, they have a gametophyte-dominant life cycle, in which cells of the plant carry only a single set of genetic information. The division name was derived from the genus name Marchantia, named by French botanist Jean Marchant after his father.

Aromatic L-amino acid decarboxylase, also known as DOPA decarboxylase (DDC), tryptophan decarboxylase, and 5-hydroxytryptophan decarboxylase, is a lyase enzyme, located in region 7p12.2-p12.1.
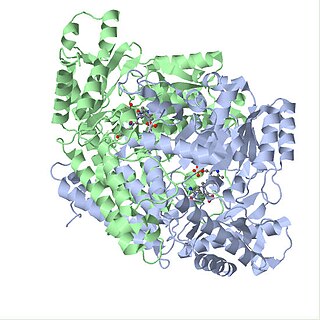
Glutamate decarboxylase or glutamic acid decarboxylase (GAD) is an enzyme that catalyzes the decarboxylation of glutamate to gamma-aminobutyric acid (GABA) and carbon dioxide. GAD uses pyridoxal-phosphate (PLP) as a cofactor. The reaction proceeds as follows:

Marchantiales is an order of thallose liverworts that includes species like Marchantia polymorpha, a widespread plant often found beside rivers, and Lunularia cruciata, a common and often troublesome weed in moist, temperate gardens and greenhouses.

Marchantiaceae is a family of liverworts in order Marchantiales. It contains a single genus Marchantia.

Lunularia is a genus of liverworts whose only species is Lunularia cruciata, the crescent-cup liverwort. Lunularia is either the only genus in the order Lunulariales, or may be placed in the order Marchantiales. The name, from Latin luna, moon, refers to the moon-shaped gemma cups.

Conocephalum is a genus of complex thalloid liverworts in the order Marchantiales and is the only extant genus in the family Conocephalaceae. Some species of Conocephalum are assigned to the Conocephalum conicum complex, which includes several cryptic species. Conocephalum species are large liverworts with distinct patterns on the upper thallus, giving the appearance of snakeskin. The species Conocephalum conicum is named for its cone-shaped reproductive structures, called archegoniophores. Common names include snakeskin liverwort, great scented liverwort and cat-tongue liverwort.

Pyruvate decarboxylase is an enzyme that catalyses the decarboxylation of pyruvic acid to acetaldehyde. It is also called 2-oxo-acid carboxylase, alpha-ketoacid carboxylase, and pyruvic decarboxylase. In anaerobic conditions, this enzyme participates in the fermentation process that occurs in yeast, especially of the genus Saccharomyces, to produce ethanol by fermentation. It is also present in some species of fish where it permits the fish to perform ethanol fermentation when oxygen is scarce. Pyruvate decarboxylase starts this process by converting pyruvate into acetaldehyde and carbon dioxide. Pyruvate decarboxylase depends on cofactors thiamine pyrophosphate (TPP) and magnesium. This enzyme should not be mistaken for the unrelated enzyme pyruvate dehydrogenase, an oxidoreductase, that catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA.
Monoicy is a sexual system in haploid plants where both sperm and eggs are produced on the same gametophyte, in contrast with dioicy, where each gametophyte produces only sperm or eggs but never both. Both monoicous and dioicous gametophytes produce gametes in gametangia by mitosis rather than meiosis, so that sperm and eggs are genetically identical with their parent gametophyte.
Carboxy-lyases, also known as decarboxylases, are carbon–carbon lyases that add or remove a carboxyl group from organic compounds. These enzymes catalyze the decarboxylation of amino acids and alpha-keto acids.

Orotidine 5′-phosphate decarboxylase or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is essential to the de novo biosynthesis of the pyrimidine nucleotides uridine triphosphate, cytidine triphosphate, and thymidine triphosphate. OMP decarboxylase has been a frequent target for scientific investigation because of its demonstrated extreme catalytic efficiency and its usefulness as a selection marker for yeast strain engineering.
Westernhope Burn Wood is a Site of Special Scientific Interest in County Durham, England. It occupies the steeply-incised ravine of the Westernhope Burn, a tributary of the River Wear, which it joins from the south about halfway between the villages of Eastgate and Westgate.

Epimartyria pardella is a species of moth belonging to the Micropterigidae family. It was described by Walsingham, Lord Thomas de Grey, in 1880. Its wingspan is 10–11 mm with a metallic brown forewing featuring three distinctive gold spots. Adults are on wing from early May to mid July and are day active. The larvae feed on liverworts, including Conocephalum conicum and Pellia species and take about two years to fully develop.

Lunularic acid is a dihydrostilbenoid found in the liverwort Lunularia cruciata and in the roots of Hydrangea macrophylla.

Conocephalum conicum, also known as the great scented liverwort or snakeskin liverwort, is a liverwort species in the genus Conocephalum. C. conicum is part of the Conocephalum conicum complex, which includes several cryptic species. The name C. conicum refers to the cone-shaped archegoniophore, which bear sporangia.

3,4′-Dihydroxystilbene is a stilbenoid found in the roots of Hydrangea macrophylla.
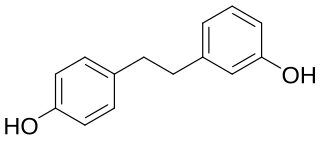
Lunularin is a dihydrostilbenoid found in common celery. It has also been found in the roots of Hydrangea macrophylla.

Conocephalum salebrosum, commonly known as snakewort, is a species of liverwort, a non-vascular land plant, with a broad, holarctic distribution. It is also known as snakeskin liverwort, cat-tongue liverwort, mushroom-headed liverwort, and great scented liverwort.
Conocephalum supradecompositum is a species of thalloid liverwort in the genus Conocephalum, of the order Marchantiales and the family Conocephalaceae. C. supradecompositum has a distribution that is mainly restricted to China and Japan. C. supradecompositum has very distinct chemical composition from the species Conocephalumconicum.














