
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

The enzyme cytochrome c oxidase or Complex IV is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.
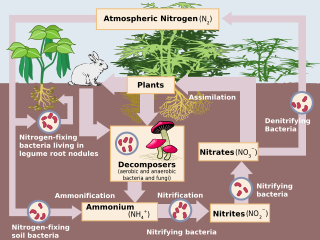
Denitrification is a microbially facilitated process where nitrate (NO3−) is reduced and ultimately produces molecular nitrogen (N2) through a series of intermediate gaseous nitrogen oxide products. Facultative anaerobic bacteria perform denitrification as a type of respiration that reduces oxidized forms of nitrogen in response to the oxidation of an electron donor such as organic matter. The preferred nitrogen electron acceptors in order of most to least thermodynamically favorable include nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O) finally resulting in the production of dinitrogen (N2) completing the nitrogen cycle. Denitrifying microbes require a very low oxygen concentration of less than 10%, as well as organic C for energy. Since denitrification can remove NO3−, reducing its leaching to groundwater, it can be strategically used to treat sewage or animal residues of high nitrogen content. Denitrification can leak N2O, which is an ozone-depleting substance and a greenhouse gas that can have a considerable influence on global warming.
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979, Trumpower's team isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein.
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.
Denitrifying bacteria are a diverse group of bacteria that encompass many different phyla. This group of bacteria, together with denitrifying fungi and archaea, is capable of performing denitrification as part of the nitrogen cycle. Denitrification is performed by a variety of denitrifying bacteria that are widely distributed in soils and sediments and that use oxidized nitrogen compounds such as nitrate and nitrite in the absence of oxygen as a terminal electron acceptor. They metabolize nitrogenous compounds using various enzymes, including nitrate reductase (NAR), nitrite reductase (NIR), nitric oxide reductase (NOR) and nitrous oxide reductase (NOS), turning nitrogen oxides back to nitrogen gas or nitrous oxide.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.
Trimethylamine N-oxide reductase is a microbial enzyme that can reduce trimethylamine N-oxide (TMAO) into trimethylamine (TMA), as part of the electron transport chain. The enzyme has been purified from E. coli and the photosynthetic bacteria Roseobacter denitrificans.
Nitric oxide reductase, an enzyme, catalyzes the reduction of nitric oxide (NO) to nitrous oxide (N2O). The enzyme participates in nitrogen metabolism and in the microbial defense against nitric oxide toxicity. The catalyzed reaction may be dependent on different participating small molecules: Cytochrome c (EC: 1.7.2.5, Nitric oxide reductase (cytochrome c)), NADPH (EC:1.7.1.14), or Menaquinone (EC:1.7.5.2).

Cytochrome c nitrite reductase (ccNiR) is a bacterial enzyme that catalyzes the six electron reduction of nitrite to ammonia; an important step in the biological nitrogen cycle. The enzyme catalyses the second step in the two step conversion of nitrate to ammonia, which allows certain bacteria to use nitrite as a terminal electron acceptor, rather than oxygen, during anaerobic conditions. During this process, ccNiR draws electrons from the quinol pool, which are ultimately provided by a dehydrogenase such as formate dehydrogenase or hydrogenase. These dehydrogenases are responsible for generating a proton motive force.
In enzymology, a nitrite reductase (NO-forming) (EC 1.7.2.1) is an enzyme that catalyzes the chemical reaction

Azurin is a small, periplasmic, bacterial blue copper protein found in Pseudomonas, Bordetella, or Alcaligenes bacteria. Azurin moderates single-electron transfer between enzymes associated with the cytochrome chain by undergoing oxidation-reduction between Cu(I) and Cu(II). Each monomer of an azurin tetramer has a molecular weight of approximately 14kDa, contains a single copper atom, is intensively blue, and has a fluorescence emission band centered at 308 nm.
Aerobic denitrification, or co-respiration, the simultaneous use of both oxygen (O2) and nitrate (NO−3) as oxidizing agents, performed by various genera of microorganisms. This process differs from anaerobic denitrification not only in its insensitivity to the presence of oxygen, but also in its higher potential to form nitrous oxide (N2O) as a byproduct.
Nitric oxide reductase (NAD(P), nitrous oxide-forming) (EC 1.7.1.14, fungal nitric oxide reductase, cytochrome P450nor, NOR (ambiguous)) is an enzyme with systematic name nitrous oxide:NAD(P) oxidoreductase. This enzyme catalyses the following chemical reaction
Nitric oxide reductase (cytochrome c) (EC 1.7.2.5) is an enzyme with systematic name nitrous oxide:ferricytochrome-c oxidoreductase. This enzyme catalyses the following chemical reaction

Urine patches in cattle pastures generate large concentrations of the greenhouse gas nitrous oxide through nitrification and denitrification processes in urine-contaminated soils. Over the past few decades, the cattle population has increased more rapidly than the human population. Between the years 2000 and 2050, the cattle population is expected to increase from 1.5 billion to 2.6 billion. When large populations of cattle are packed into pastures, excessive amounts of urine soak into soils. This increases the rate at which nitrification and denitrification occur and produce nitrous oxide. Currently, nitrous oxide is one of the single most important ozone-depleting emissions and is expected to remain the largest throughout the 21st century.
Dissimilatory nitrate reduction to ammonium (DNRA), also known as nitrate/nitrite ammonification, is the result of anaerobic respiration by chemoorganoheterotrophic microbes using nitrate (NO3−) as an electron acceptor for respiration. In anaerobic conditions microbes which undertake DNRA oxidise organic matter and use nitrate (rather than oxygen) as an electron acceptor, reducing it to nitrite, and then to ammonium (NO3− → NO2− → NH4+).









