
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.
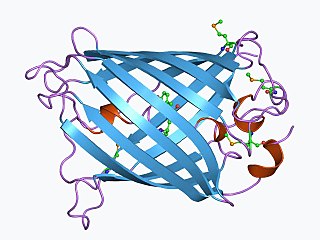
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria and is sometimes called avGFP. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

In molecular biology and biotechnology, a fluorescent tag, also known as a fluorescent label or fluorescent probe, is a molecule that is attached chemically to aid in the detection of a biomolecule such as a protein, antibody, or amino acid. Generally, fluorescent tagging, or labeling, uses a reactive derivative of a fluorescent molecule known as a fluorophore. The fluorophore selectively binds to a specific region or functional group on the target molecule and can be attached chemically or biologically. Various labeling techniques such as enzymatic labeling, protein labeling, and genetic labeling are widely utilized. Ethidium bromide, fluorescein and green fluorescent protein are common tags. The most commonly labelled molecules are antibodies, proteins, amino acids and peptides which are then used as specific probes for detection of a particular target.

A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large macromolecules such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as primary metabolites, secondary metabolites and natural products. A more general name for this class of material is biological materials. Biomolecules are an important element of living organisms, those biomolecules are often endogenous, produced within the organism but organisms usually need exogenous biomolecules, for example certain nutrients, to survive.
A fluorophore is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds.

The plasma membranes of cells contain combinations of glycosphingolipids, cholesterol and protein receptors organised in glycolipoprotein lipid microdomains termed lipid rafts. Their existence in cellular membranes remains somewhat controversial. It has been proposed that they are specialized membrane microdomains which compartmentalize cellular processes by serving as organising centers for the assembly of signaling molecules, allowing a closer interaction of protein receptors and their effectors to promote kinetically favorable interactions necessary for the signal transduction. Lipid rafts influence membrane fluidity and membrane protein trafficking, thereby regulating neurotransmission and receptor trafficking. Lipid rafts are more ordered and tightly packed than the surrounding bilayer, but float freely within the membrane bilayer. Although more common in the cell membrane, lipid rafts have also been reported in other parts of the cell, such as the Golgi apparatus and lysosomes.

A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image.
Phosphatidic acids are anionic phospholipids important to cell signaling and direct activation of lipid-gated ion channels. Hydrolysis of phosphatidic acid gives rise to one molecule each of glycerol and phosphoric acid and two molecules of fatty acids. They constitute about 0.25% of phospholipids in the bilayer.

Spatiotemporal gene expression is the activation of genes within specific tissues of an organism at specific times during development. Gene activation patterns vary widely in complexity. Some are straightforward and static, such as the pattern of tubulin, which is expressed in all cells at all times in life. Some, on the other hand, are extraordinarily intricate and difficult to predict and model, with expression fluctuating wildly from minute to minute or from cell to cell. Spatiotemporal variation plays a key role in generating the diversity of cell types found in developed organisms; since the identity of a cell is specified by the collection of genes actively expressed within that cell, if gene expression was uniform spatially and temporally, there could be at most one kind of cell.

Ceramides are a family of waxy lipid molecules. A ceramide is composed of sphingosine and a fatty acid. Ceramides are found in high concentrations within the cell membrane of eukaryotic cells, since they are component lipids that make up sphingomyelin, one of the major lipids in the lipid bilayer. Contrary to previous assumptions that ceramides and other sphingolipids found in cell membrane were purely supporting structural elements, ceramide can participate in a variety of cellular signaling: examples include regulating differentiation, proliferation, and programmed cell death (PCD) of cells.
In biology, cell signaling or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. It is a fundamental property of all cells in every living organism such as bacteria, plants, and animals. Signals that originate from outside a cell can be physical agents like mechanical pressure, voltage, temperature, light, or chemical signals. Chemical signals can be hydrophobic or hydrophillic. Cell signaling can occur over short or long distances, and as a result can be classified as autocrine, juxtacrine, intracrine, paracrine, or endocrine. Signaling molecules can be synthesized from various biosynthetic pathways and released through passive or active transports, or even from cell damage.

Lipid signaling, broadly defined, refers to any biological signaling event involving a lipid messenger that binds a protein target, such as a receptor, kinase or phosphatase, which in turn mediate the effects of these lipids on specific cellular responses. Lipid signaling is thought to be qualitatively different from other classical signaling paradigms because lipids can freely diffuse through membranes One consequence of this is that lipid messengers cannot be stored in vesicles prior to release and so are often biosynthesized "on demand" at their intended site of action. As such, many lipid signaling molecules cannot circulate freely in solution but, rather, exist bound to special carrier proteins in serum.

Protein kinase C beta type is an enzyme that in humans is encoded by the PRKCB gene.

Mitogen-activated protein kinase 7 also known as MAP kinase 7 is an enzyme that in humans is encoded by the MAPK7 gene.
EosFP is a photoactivatable green to red fluorescent protein. Its green fluorescence (516 nm) switches to red (581 nm) upon UV irradiation of ~390 nm due to a photo-induced modification resulting from a break in the peptide backbone near the chromophore. Eos was first discovered as a tetrameric protein in the stony coral Lobophyllia hemprichii. Like other fluorescent proteins, Eos allows for applications such as the tracking of fusion proteins, multicolour labelling and tracking of cell movement. Several variants of Eos have been engineered for use in specific study systems including mEos2, mEos4 and CaMPARI.
mCherry is a member of the mFruits family of monomeric red fluorescent proteins (mRFPs). As a RFP, mCherry was derived from DsRed of Discosoma sea anemones unlike green fluorescent proteins (GFPs) which are often derived from Aequoera victoria jellyfish. Fluorescent proteins are used to tag components in the cell, so they can be studied using fluorescence spectroscopy and fluorescence microscopy. mCherry absorbs light between 540-590 nm and emits light in the range of 550-650 nm. mCherry belongs to the group of fluorescent protein chromophores used as instruments to visualize genes and analyze their functions in experiments. Genome editing has been improved greatly through the precise insertion of these fluorescent protein tags into the genetic material of many diverse organisms. Most comparisons between the brightness and photostability of different fluorescent proteins have been made in vitro, removed from biological variables that affect protein performance in cells or organisms. It is hard to perfectly simulate cellular environments in vitro, and the difference in environment could have an effect on the brightness and photostability.
Photo-activated localization microscopy and stochastic optical reconstruction microscopy (STORM) are widefield fluorescence microscopy imaging methods that allow obtaining images with a resolution beyond the diffraction limit. The methods were proposed in 2006 in the wake of a general emergence of optical super-resolution microscopy methods, and were featured as Methods of the Year for 2008 by the Nature Methods journal. The development of PALM as a targeted biophysical imaging method was largely prompted by the discovery of new species and the engineering of mutants of fluorescent proteins displaying a controllable photochromism, such as photo-activatible GFP. However, the concomitant development of STORM, sharing the same fundamental principle, originally made use of paired cyanine dyes. One molecule of the pair, when excited near its absorption maximum, serves to reactivate the other molecule to the fluorescent state.
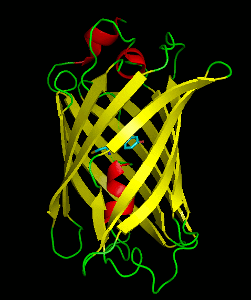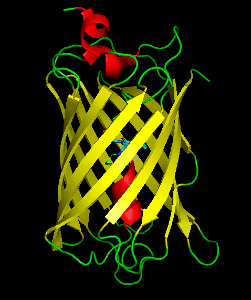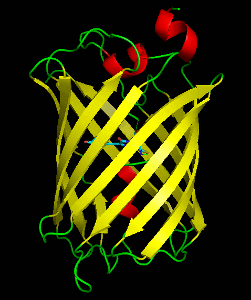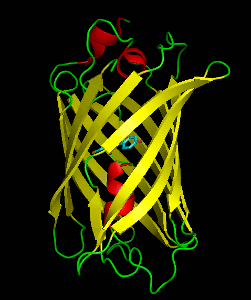
Dronpa is a reversibly switchable photoactivatable fluorescent protein that is 2.5 times as bright as EGFP. Dronpa gets switched off by strong illumination with 488 nm (blue) light and this can be reversed by weak 405 nm UV light. A single dronpa molecule can be switched on and off over 100 times. It has an excitation peak at 503 nm and an emission peak at 518 nm.













