
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

A cyclic nucleotide (cNMP) is a single-phosphate nucleotide with a cyclic bond arrangement between the sugar and phosphate groups. Like other nucleotides, cyclic nucleotides are composed of three functional groups: a sugar, a nitrogenous base, and a single phosphate group. As can be seen in the cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) images, the 'cyclic' portion consists of two bonds between the phosphate group and the 3' and 5' hydroxyl groups of the sugar, very often a ribose.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
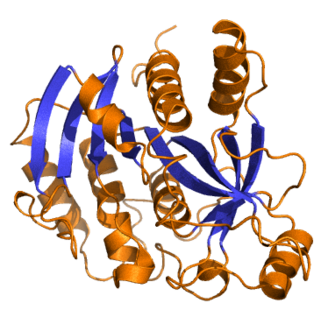
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.
In enzymology, an allyl-alcohol dehydrogenase (EC 1.1.1.54) is an enzyme that catalyzes the chemical reaction
In enzymology, a tryptophan 2-monooxygenase (EC 1.13.12.3) is an enzyme that catalyzes the chemical reaction

ADP-ribose diphosphatase (EC 3.6.1.13) is an enzyme that catalyzes a hydrolysis reaction in which water nucleophilically attacks ADP-ribose to produce AMP and D-ribose 5-phosphate. Enzyme hydrolysis occurs by the breakage of a phosphoanhydride bond and is dependent on Mg2+ ions that are held in complex by the enzyme.

In enzymology, a nucleoside-diphosphatase (EC 3.6.1.6) is an enzyme that catalyzes the chemical reaction
The enzyme feruloyl esterase (EC 3.1.1.73) catalyzes the reaction
In enzymology, a NMN nucleosidase (EC 3.2.2.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a purine nucleosidase (EC 3.2.2.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a ribosylpyrimidine nucleosidase (EC 3.2.2.8) is an enzyme that catalyzes the chemical reaction
In enzymology, an adenosine-phosphate deaminase (EC 3.5.4.17) is an enzyme that catalyzes the chemical reaction
2-Formylbenzoate dehydrogenase (EC 1.2.1.78, 2-carboxybenzaldehyde dehydrogenase, 2CBAL dehydrogenase, PhdK) is an enzyme with systematic name 2-formylbenzoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Mn2+-dependent ADP-ribose/CDP-alcohol diphosphatase (EC 3.6.1.53, Mn2+-dependent ADP-ribose/CDP-alcohol pyrophosphatase, ADPRibase-Mn) is an enzyme with systematic name CDP-choline phosphohydrolase. This enzyme catalyses the following chemical reaction
Aminopyrimidine aminohydrolase (EC 3.5.99.2, thiaminase, thiaminase II, tenA (gene)) is an enzyme with systematic name 4-amino-5-aminomethyl-2-methylpyrimidine aminohydrolase. This enzyme catalyses the following chemical reaction
Arthrobacter luteus (ALU) is a species of gram-positive bacteria in the genus Arthrobacter. A. luteus is facultatively anaerobic, pleomorphic, branching, non-motile, non-sporulating, non-acid-fast, catalase-positive, and rod-shaped.








