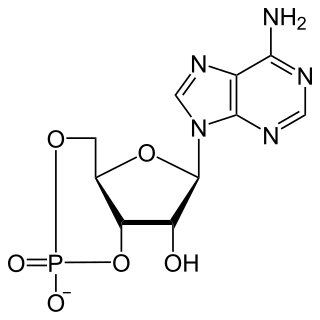
Cyclic adenosine monophosphate is a second messenger, or cellular signal occurring within cells, that is important in many biological processes. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms, conveying the cAMP-dependent pathway.

A cyclic nucleotide (cNMP) is a single-phosphate nucleotide with a cyclic bond arrangement between the sugar and phosphate groups. Like other nucleotides, cyclic nucleotides are composed of three functional groups: a sugar, a nitrogenous base, and a single phosphate group. As can be seen in the cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) images, the 'cyclic' portion consists of two bonds between the phosphate group and the 3' and 5' hydroxyl groups of the sugar, very often a ribose.

Nucleotide exchange factors (NEFs) are proteins that stimulate the exchange (replacement) of nucleoside diphosphates for nucleoside triphosphates bound to other proteins.

CAMP responsive element binding protein 1, also known as CREB-1, is a protein that in humans is encoded by the CREB1 gene. This protein binds the cAMP response element, a DNA nucleotide sequence present in many viral and cellular promoters. The binding of CREB1 stimulates transcription.

Cyclic AMP-dependent transcription factor ATF-1 is a protein that in humans is encoded by the ATF1 gene.

Ras-related protein Rap-2a is a protein that in humans is encoded by the RAP2A gene. RAP2A is a member of the Ras-related protein family.

Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated ion channel 2 is a protein that in humans is encoded by the HCN2 gene.
Rap guanine nucleotide exchange factor (GEF) 4 (RAPGEF4), also known as exchange protein directly activated by cAMP 2 (EPAC2) is a protein that in humans is encoded by the RAPGEF4 gene.

Ras-related protein Rap-2b is a protein that in humans is encoded by the RAP2B gene. RAP2B belongs to the Ras-related protein family.

Mitochondrial GTPase 1 is an enzyme that in humans is encoded by the MTG1 gene.

Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 is a protein that in humans is encoded by the HCN1 gene.
Hyperpolarization-activated cyclic nucleotide–gated (HCN) channels are integral membrane proteins that serve as nonselective voltage-gated cation channels in the plasma membranes of heart and brain cells. HCN channels are sometimes referred to as pacemaker channels because they help to generate rhythmic activity within groups of heart and brain cells. HCN channels are activated by membrane hyperpolarization, are permeable to Na + and K +, and are constitutively open at voltages near the resting membrane potential. HCN channels are encoded by four genes and are widely expressed throughout the heart and the central nervous system.

Rap guanine nucleotide exchange factor 2 is a protein that in humans is encoded by the RAPGEF2 gene.

RAS guanyl-releasing protein 2 is a protein that in humans is encoded by the RASGRP2 gene.

Cyclic nucleotide gated channel alpha 2, also known as CNGA2, is a human gene encoding an ion channel protein.

Rap1 GTPase-GDP dissociation stimulator 1 is an enzyme that in humans is encoded by the RAP1GDS1 gene.

Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 is a protein that in humans is encoded by the HCN3 gene.

Rap guanine nucleotide exchange factor 5 is a protein that in humans is encoded by the RAPGEF5 gene.
In the field of molecular biology, the cAMP-dependent pathway, also known as the adenylyl cyclase pathway, is a G protein-coupled receptor-triggered signaling cascade used in cell communication.

Cyclic di-AMP is a second messenger used in signal transduction in bacteria and archaea. It is present in many Gram-positive bacteria, some Gram-negative species, and archaea of the phylum Euryarchaeota.





















