
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

A prophage is a bacteriophage genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors.

Transduction is the process by which foreign DNA is introduced into a cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.

The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
Microviridae is a family of bacteriophages with a single-stranded DNA genome. The name of this family is derived from the ancient Greek word μικρός (mikrós), meaning "small". This refers to the size of their genomes, which are among the smallest of the DNA viruses. Enterobacteria, intracellular parasitic bacteria, and spiroplasma serve as natural hosts. There are 22 species in this family, divided among seven genera and two subfamilies.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.
Escherichia virus HK97, often shortened to HK97, is a species of virus that infects Escherichia coli and related bacteria. It is named after Hong Kong (HK), where it was first located. HK97 has a double-stranded DNA genome.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.

Escherichia virus T5, sometimes called Bacteriophage T5 is a caudal virus within the family Demerecviridae. This bacteriophage specifically infects E. coli bacterial cells and follows a lytic life cycle.

Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus Peduovirus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.
A prohead or procapsid is an immature viral capsid structure formed in the early stages of self-assembly of some bacteriophages, including the Caudovirales or tailed bacteriophages. Production and assembly of stable proheads is an essential precursor to bacteriophage genome packaging; this packaging activity can be replicated in vitro. The prohead structure may take a different shape from the head of a mature virion, as seen with the prohead of Bacillus subtilis phage φ29.

Spiroplasma phage 1-R8A2B is a filamentous bacteriophage in the genus Vespertiliovirus of the family Plectroviridae, part of the group of single-stranded DNA viruses. The virus has many synonyms, such as SpV1-R8A2 B, Spiroplasma phage 1, and Spiroplasma virus 1, SpV1. SpV1-R8A2 B infects Spiroplasma citri. Its host itself is a prokaryotic pathogen for citrus plants, causing Citrus stubborn disease.

Teseptimavirus is a genus of viruses in the order Caudovirales, in the family Autographiviridae, in the subfamily Studiervirinae. Bacteria serve as the natural host, with transmission achieved through passive diffusion. There are currently 17 species in this genus, including the type species Escherichia virus T7.
Lederbergvirus is a genus of viruses in the order Caudovirales, in the family Podoviridae. Bacteria serve as natural hosts, with transmission achieved through passive diffusion. There are six species in this genus.
Escherichia virus CC31, formerly known as Enterobacter virus CC31, is a dsDNA bacteriophage of the subfamily Tevenvirinae responsible for infecting the bacteria family of Enterobacteriaceae. It is one of two discovered viruses of the genus Karamvirus, diverging away from the previously discovered T4virus, as a clonal complex (CC). CC31 was first isolated from Escherichia coli B strain S/6/4 and is primarily associated with Escherichia, even though is named after Enterobacter.
Arbitrium is a viral peptide produced by bacteriophages to communicate with each other and decide host cell fate. It is six amino acids(aa) long, and so is also referred to as a hexapeptide. It is produced when a phage infects a bacterial host. and signals to other phages that the host has been infected.

Duplodnaviria is a realm of viruses that includes all double-stranded DNA viruses that encode the HK97 fold major capsid protein. The HK97 fold major capsid protein is the primary component of the viral capsid, which stores the viral deoxyribonucleic acid (DNA). Viruses in the realm also share a number of other characteristics, such as an icosahedral capsid, an opening in the viral capsid called a portal, a protease enzyme that empties the inside of the capsid prior to DNA packaging, and a terminase enzyme that packages viral DNA into the capsid.
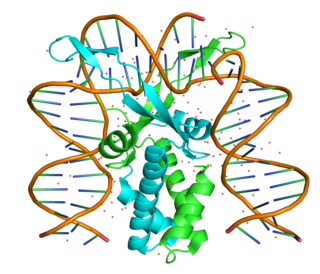
The integration host factor (IHF) is a bacterial DNA binding protein complex that facilitates genetic recombination, replication, and transcription by binding to specific DNA sequences and bending the DNA. It also facilitates the integration of foreign DNA into the host genome. It is a heterodimeric complex composed of two homologous subunits IHFalpha and IHFbeta.














