
Cyanobacteria, also called Cyanobacteriota or Cyanophyta, are a phylum of gram-negative bacteria that obtain energy via photosynthesis. The name cyanobacteria refers to their color, which similarly forms the basis of cyanobacteria's common name, blue-green algae, although they are not usually scientifically classified as algae. They appear to have originated in a freshwater or terrestrial environment. Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria.

Synechococcus is a unicellular cyanobacterium that is very widespread in the marine environment. Its size varies from 0.8 to 1.5 µm. The photosynthetic coccoid cells are preferentially found in well–lit surface waters where it can be very abundant. Many freshwater species of Synechococcus have also been described.

Cyanophages are viruses that infect cyanobacteria, also known as Cyanophyta or blue-green algae. Cyanobacteria are a phylum of bacteria that obtain their energy through the process of photosynthesis. Although cyanobacteria metabolize photoautotrophically like eukaryotic plants, they have prokaryotic cell structure. Cyanophages can be found in both freshwater and marine environments. Marine and freshwater cyanophages have icosahedral heads, which contain double-stranded DNA, attached to a tail by connector proteins. The size of the head and tail vary among species of cyanophages. Cyanophages infect a wide range of cyanobacteria and are key regulators of the cyanobacterial populations in aquatic environments, and may aid in the prevention of cyanobacterial blooms in freshwater and marine ecosystems. These blooms can pose a danger to humans and other animals, particularly in eutrophic freshwater lakes. Infection by these viruses is highly prevalent in cells belonging to Synechococcus spp. in marine environments, where up to 5% of cells belonging to marine cyanobacterial cells have been reported to contain mature phage particles.
Gloeobacter is a genus of cyanobacteria. It is the sister group to all other cyanobacteria. Gloeobacter is unique among cyanobacteria in not having thylakoids, which are characteristic for all other cyanobacteria and chloroplasts. Instead, the light-harvesting complexes, that consist of different proteins, sit on the inside of the plasma membrane among the (cytoplasm). Subsequently, the proton gradient in Gloeobacter is created over the plasma membrane, where it forms over the thylakoid membrane in cyanobacteria and chloroplasts.
Synechocystis sp. PCC6803 is a strain of unicellular, freshwater cyanobacteria. Synechocystis sp. PCC6803 is capable of both phototrophic growth by oxygenic photosynthesis during light periods and heterotrophic growth by glycolysis and oxidative phosphorylation during dark periods. Gene expression is regulated by a circadian clock and the organism can effectively anticipate transitions between the light and dark phases.
Bacterial circadian rhythms, like other circadian rhythms, are endogenous "biological clocks" that have the following three characteristics: (a) in constant conditions they oscillate with a period that is close to, but not exactly, 24 hours in duration, (b) this "free-running" rhythm is temperature compensated, and (c) the rhythm will entrain to an appropriate environmental cycle.
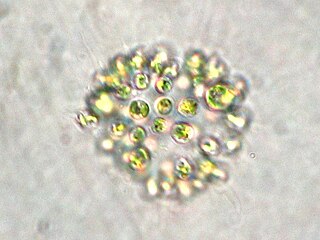
Microcystis aeruginosa is a species of freshwater cyanobacteria that can form harmful algal blooms of economic and ecological importance. They are the most common toxic cyanobacterial bloom in eutrophic fresh water. Cyanobacteria produce neurotoxins and peptide hepatotoxins, such as microcystin and cyanopeptolin. Microcystis aeruginosa produces numerous congeners of microcystin, with microcystin-LR being the most common. Microcystis blooms have been reported in at least 108 countries, with the production of microcystin noted in at least 79.

In molecular biology, the cyanobacterial clock proteins are the main circadian regulator in cyanobacteria. The cyanobacterial clock proteins comprise three proteins: KaiA, KaiB and KaiC. The kaiABC complex may act as a promoter-nonspecific transcription regulator that represses transcription, possibly by acting on the state of chromosome compaction. This complex is expressed from a KaiABC operon.
In molecular biology, Cyanobacterial non-coding RNAs are non-coding RNAs which have been identified in species of cyanobacteria. Large scale screens have identified 21 Yfr in the marine cyanobacterium Prochlorococcus and related species such as Synechococcus. These include the Yfr1 and Yfr2 RNAs. In Prochlorococcus and Synechocystis, non-coding RNAs have been shown to regulate gene expression. NsiR4, widely conserved throughout the cyanobacterial phylum, has been shown to be involved in nitrogen assimilation control in Synechocystis sp. PCC 6803 and in the filamentous, nitrogen-fixing Anabaena sp. PCC 7120.

KaiC is a gene belonging to the KaiABC gene cluster that, together, regulate bacterial circadian rhythms, specifically in cyanobacteria. KaiC encodes for the KaiC protein, which interacts with the KaiA and KaiB proteins in a post-translational oscillator (PTO). The PTO is cyanobacteria master clock that is controlled by sequences of phosphorylation of KaiC protein. Regulation of KaiABC expression and KaiABC phosphorylation is essential for cyanobacteria circadian rhythmicity, and is particularly important for regulating cyanobacteria processes such as nitrogen fixation, photosynthesis, and cell division. Studies have shown similarities to Drosophila, Neurospora, and mammalian clock models in that the kaiABC regulation of the cyanobacteria slave circadian clock is also based on a transcription translation feedback loop (TTFL). KaiC protein has both auto-kinase and auto-phosphatase activity and functions as the circadian regulator in both the PTO and the TTFL. KaiC has been found to not only suppress kaiBC when overexpressed, but also suppress circadian expression of all genes in the cyanobacterial genome.

Cyanophycinase (EC 3.4.15.6, cyanophycin degrading enzyme, beta-Asp-Arg hydrolysing enzyme, CGPase, CphB, CphE, cyanophycin granule polypeptidase, extracellular CGPase) is an enzyme. It catalyses the following chemical reaction
Lycopene β-cyclase is an enzyme with systematic name carotenoid beta-end group lyase (decyclizing). This enzyme catalyses the following chemical reaction

Cyanothece is a genus of unicellular, diazotrophic, oxygenic photosynthesizing cyanobacteria.
kaiA is a gene in the "kaiABC" gene cluster that plays a crucial role in the regulation of bacterial circadian rhythms, such as in the cyanobacterium Synechococcus elongatus. For these bacteria, regulation of kaiA expression is critical for circadian rhythm, which determines the twenty-four-hour biological rhythm. In addition, KaiA functions with a negative feedback loop in relation with kaiB and KaiC. The kaiA gene makes KaiA protein that enhances phosphorylation of KaiC while KaiB inhibits activity of KaiA.
Myxoxanthophyll is a carotenoid glycoside pigment present in the photosynthetic apparatus of cyanobacteria. It is named after the word "Myxophyceae", a former term for cyanobacteria. As a monocyclic xanthophyll, it has a yellowish color. It is required for normal cell wall structure and thylakoid organization in the cyanobacterium Synechocystis. The pigment is unusual because it is glycosylated on the 2'-OH rather than the 1'-OH position of the molecule. Myxoxanthophyll was first isolated from Oscillatoria rubenscens in 1936.
KaiB is a gene located in the highly-conserved kaiABC gene cluster of various cyanobacterial species. Along with KaiA and KaiC, KaiB plays a central role in operation of the cyanobacterial circadian clock. Discovery of the Kai genes marked the first-ever identification of a circadian oscillator in a prokaryotic species. Moreover, characterization of the cyanobacterial clock demonstrated the existence of transcription-independent, post-translational mechanisms of rhythm generation, challenging the universality of the transcription-translation feedback loop model of circadian rhythmicity.
Susan Golden is a Professor of molecular biology known for her research in circadian rhythms. She is currently a faculty member at UC San Diego.

Oscillatoria brevis, is a species of the genus Oscillatoria first identified in 1892. It is a blue-green filamentous cyanobacterium, which can be found in brackish and fresh waterways. O. brevis can also be isolated from soil.
A plastid is a membrane-bound organelle found in plants, algae and other eukaryotic organisms that contribute to the production of pigment molecules. Most plastids are photosynthetic, thus leading to color production and energy storage or production. There are many types of plastids in plants alone, but all plastids can be separated based on the number of times they have undergone endosymbiotic events. Currently there are three types of plastids; primary, secondary and tertiary. Endosymbiosis is reputed to have led to the evolution of eukaryotic organisms today, although the timeline is highly debated.

Synechocystis is a genus of unicellular, freshwater cyanobacteria in the family Merismopediaceae. It includes a strain, Synechocystis sp. PCC 6803, which is a well studied model organism.










