Related Research Articles

In a dynamical system, bistability means the system has two stable equilibrium states. Something that is bistable can be resting in either of two states. An example of a mechanical device which is bistable is a light switch. The switch lever is designed to rest in the "on" or "off" position, but not between the two. Bistable behavior can occur in mechanical linkages, electronic circuits, nonlinear optical systems, chemical reactions, and physiological and biological systems.

In genetics, an enhancer is a short region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes.
A circadian clock, or circadian oscillator, is a biochemical oscillator that cycles with a stable phase and is synchronized with solar time.

Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a protein complex that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens. NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory.
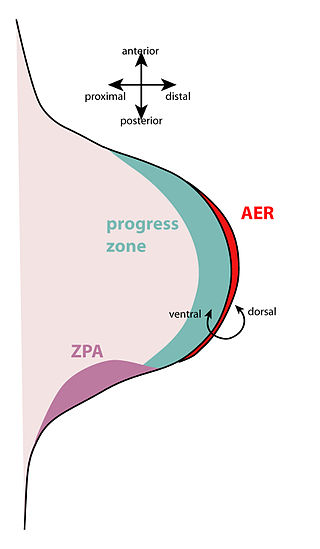
The apical ectodermal ridge (AER) is a structure that forms from the ectodermal cells at the distal end of each limb bud and acts as a major signaling center to ensure proper development of a limb. After the limb bud induces AER formation, the AER and limb mesenchyme—including the zone of polarizing activity (ZPA)—continue to communicate with each other to direct further limb development.
Limb development in vertebrates is an area of active research in both developmental and evolutionary biology, with much of the latter work focused on the transition from fin to limb.
The limb bud is a structure formed early in vertebrate limb development. As a result of interactions between the ectoderm and underlying mesoderm, formation occurs roughly around the fourth week of development. In the development of the human embryo the upper limb bud appears in the third week and the lower limb bud appears four days later.

CLOCK is a gene encoding a basic helix-loop-helix-PAS transcription factor that is known to affect both the persistence and period of circadian rhythms.
Gremlin is an inhibitor in the TGF beta signaling pathway. It primarily inhibits bone morphogenesis and is implicated in disorders of increased bone formation and several cancers.

Neuronal PAS domain protein 2 (NPAS2) also known as member of PAS protein 4 (MOP4) is a transcription factor protein that in humans is encoded by the NPAS2 gene. NPAS2 is paralogous to CLOCK, and both are key proteins involved in the maintenance of circadian rhythms in mammals. In the brain, NPAS2 functions as a generator and maintainer of mammalian circadian rhythms. More specifically, NPAS2 is an activator of transcription and translation of core clock and clock-controlled genes through its role in a negative feedback loop in the suprachiasmatic nucleus (SCN), the brain region responsible for the control of circadian rhythms.

FBXL3 is a gene in humans and mice that encodes the F-box/LRR-repeat protein 3 (FBXL3). FBXL3 is a member of the F-box protein family, which constitutes one of the four subunits in the SCF ubiquitin ligase complex.
The IκB kinase is an enzyme complex that is involved in propagating the cellular response to inflammation.

Transcription factor p65 also known as nuclear factor NF-kappa-B p65 subunit is a protein that in humans is encoded by the RELA gene.

RAR-related orphan receptor alpha (RORα), also known as NR1F1 is a nuclear receptor that in humans is encoded by the RORA gene. RORα participates in the transcriptional regulation of some genes involved in circadian rhythm. In mice, RORα is essential for development of cerebellum through direct regulation of genes expressed in Purkinje cells. It also plays an essential role in the development of type 2 innate lymphoid cells (ILC2) and mutant animals are ILC2 deficient. In addition, although present in normal numbers, the ILC3 and Th17 cells from RORα deficient mice are defective for cytokine production.

Inhibitor of nuclear factor kappa-B kinase subunit alpha (IKK-α) also known as IKK1 or conserved helix-loop-helix ubiquitous kinase (CHUK) is a protein kinase that in humans is encoded by the CHUK gene. IKK-α is part of the IκB kinase complex that plays an important role in regulating the NF-κB transcription factor. However, IKK-α has many additional cellular targets, and is thought to function independently of the NF-κB pathway to regulate epidermal differentiation.

The zone of polarizing activity (ZPA) is an area of mesenchyme that contains signals which instruct the developing limb bud to form along the anterior/posterior axis. Limb bud is undifferentiated mesenchyme enclosed by an ectoderm covering. Eventually, the limb bud develops into bones, tendons, muscles and joints. Limb bud development relies not only on the ZPA, but also many different genes, signals, and a unique region of ectoderm called the apical ectodermal ridge (AER). Research by Saunders and Gasseling in 1948 identified the AER and its subsequent involvement in proximal distal outgrowth. Twenty years later, the same group did transplantation studies in chick limb bud and identified the ZPA. It wasn't until 1993 that Todt and Fallon showed that the AER and ZPA are dependent on each other.
Aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL) or brain and muscle ARNT-Like 1 (BMAL1) is a protein that in humans is encoded by the ARNTL gene on chromosome 11, region p15.3. It's also known as BMAL1, MOP3, and, less commonly, bHLHe5, BMAL, BMAL1C, JAP3, PASD3, and TIC.
In molecular biology, an oscillating gene is a gene that is expressed in a rhythmic pattern or in periodic cycles. Oscillating genes are usually circadian and can be identified by periodic changes in the state of an organism. Circadian rhythms, controlled by oscillating genes, have a period of approximately 24 hours. For example, plant leaves opening and closing at different times of the day or the sleep-wake schedule of animals can all include circadian rhythms. Other periods are also possible, such as 29.5 days resulting from circalunar rhythms or 12.4 hours resulting from circatidal rhythms. Oscillating genes include both core clock component genes and output genes. A core clock component gene is a gene necessary for to the pacemaker. However, an output oscillating gene, such as the AVP gene, is rhythmic but not necessary to the pacemaker.
A series of biochemical switches control transitions between and within the various phases of the cell cycle. The cell cycle is a series of complex, ordered, sequential events that control how a single cell divides into two cells, and involves several different phases. The phases include the G1 and G2 phases, DNA replication or S phase, and the actual process of cell division, mitosis or M phase. During the M phase, the chromosomes separate and cytokinesis occurs.
Transcription-translation feedback loop (TTFL) is a cellular model for explaining circadian rhythms in behavior and physiology. Widely conserved across species, the TTFL is auto-regulatory, in which transcription of clock genes is regulated by their own protein products.
References
- 1 2 Brandman, Onn; James E. Ferrell Jr; Rong Li; Tobias Meyer (2005). "Interlinked Fast and Slow Positive Feedback Loops Drive Reliable Cell Decisions". Science. 310 (5747): 496–498. Bibcode:2005Sci...310..496B. doi:10.1126/science.1113834. PMC 3175767 . PMID 16239477.
- ↑ Zhang, Xiao-Peng; Zhang Cheng; Feng Liu; Wei Wang (2007). "Linking fast and slow positive feedback loops creates an optimal bistable switch in cell signaling". Phys. Rev. E. 76 (3): 031924. Bibcode:2007PhRvE..76c1924Z. doi:10.1103/physreve.76.031924. PMID 17930288.
- ↑ Smolen, Paul; Douglas A. Baxter; John H. Byrne (2009). "Interlinked dual-time feedback loops can enhance robustness to stochasticity and persistence of memory". Phys. Rev. E. 79 (3): 031902. arXiv: 1208.6050 . Bibcode:2009PhRvE..79c1902S. doi:10.1103/physreve.79.031902. PMC 2742492 . PMID 19391966.
- ↑ Hoffmann, Alexander; Andre Levchenko; Martin L. Scott; David Baltimore (2002). "The I B–NF- B Signaling Module: Temporal Control and Selective Gene Activation". Science. 298 (5596): 1241–1245. Bibcode:2002Sci...298.1241H. doi:10.1126/science.1071914. PMID 12424381. S2CID 11834415.
- 1 2 Zeller, Rolf; Javier López-Ríos; Aimée Zuniga (2009). "Vertebrate limb bud development: moving towards integrative analysis of organogenesis". Nature Reviews Genetics. 10 (12): 845–858. doi:10.1038/nrg2681. PMID 19920852. S2CID 31202624.
- ↑ Bénazet, Jean-Denis; Mirko Bischofberger; Eva Tiecke; Alexandre Gonçalves; James Martin; Aimée Zuniga; Felix Naef; Rolf Zeller (2009). "A Self-Regulatory System of Interlinked Signaling Feedback Loops Controls Mouse Limb Patterning". Science. 323 (5917): 1050–3. Bibcode:2009Sci...323.1050B. doi:10.1126/science.1168755. PMID 19229034. S2CID 25309127.
- ↑ Petrillo, Ezequiel; Sabrina E. Sanchez; Alberto R. Kornblihtt; Marcelo J. Yanovsky (2011). "Alternative Splicing Adds a New Loop to the Circadian Clock" (PDF). Communicative & Integrative Biology. 4:2.