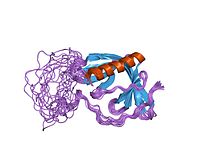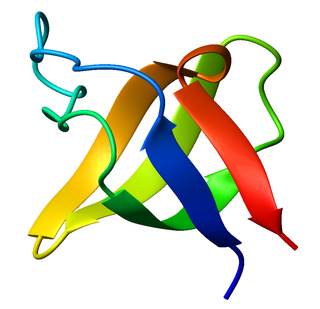
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.

Microfilaments, also called actin filaments, are protein filaments in the cytoplasm of eukaryotic cells that form part of the cytoskeleton. They are primarily composed of polymers of actin, but are modified by and interact with numerous other proteins in the cell. Microfilaments are usually about 7 nm in diameter and made up of two strands of actin. Microfilament functions include cytokinesis, amoeboid movement, cell motility, changes in cell shape, endocytosis and exocytosis, cell contractility, and mechanical stability. Microfilaments are flexible and relatively strong, resisting buckling by multi-piconewton compressive forces and filament fracture by nanonewton tensile forces. In inducing cell motility, one end of the actin filament elongates while the other end contracts, presumably by myosin II molecular motors. Additionally, they function as part of actomyosin-driven contractile molecular motors, wherein the thin filaments serve as tensile platforms for myosin's ATP-dependent pulling action in muscle contraction and pseudopod advancement. Microfilaments have a tough, flexible framework which helps the cell in movement.

The Wiskott–Aldrich syndrome protein (WASp) is a 502-amino acid protein expressed in cells of the hematopoietic system that in humans is encoded by the WAS gene. In the inactive state, WASp exists in an autoinhibited conformation with sequences near its C-terminus binding to a region near its N-terminus. Its activation is dependent upon CDC42 and PIP2 acting to disrupt this interaction, causing the WASp protein to 'open'. This exposes a domain near the WASp C-terminus that binds to and activates the Arp2/3 complex. Activated Arp2/3 nucleates new F-actin.
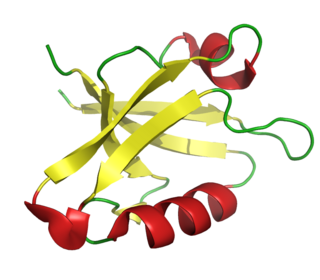
The PDZ domain is a common structural domain of 80-90 amino-acids found in the signaling proteins of bacteria, yeast, plants, viruses and animals. Proteins containing PDZ domains play a key role in anchoring receptor proteins in the membrane to cytoskeletal components. Proteins with these domains help hold together and organize signaling complexes at cellular membranes. These domains play a key role in the formation and function of signal transduction complexes. PDZ domains also play a highly significant role in the anchoring of cell surface receptors to the actin cytoskeleton via mediators like NHERF and ezrin.

Testin is a protein that in humans is encoded by the TES gene located on chromosome 7. TES is a 47 kDa protein composed of 421 amino acids found at focal adhesions and is thought to have a role in regulation of cell motility. In addition to this, TES functions as a tumour suppressor. The TES gene is located within a fragile region of chromosome 7, and the promoter elements of the TES gene have been shown to be susceptible to methylation – this prevents the expression of the TES protein. TES came to greater prominence towards the end of 2007 as a potential mechanism for its tumour suppressor function was published.
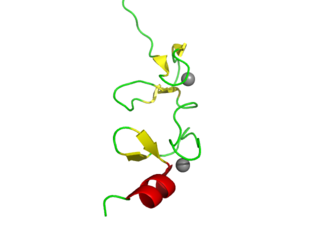
LIM domains are protein structural domains, composed of two contiguous zinc fingers, separated by a two-amino acid residue hydrophobic linker. The domain name is an acronym of the three genes in which it was first identified. LIM is a protein interaction domain that is involved in binding to many structurally and functionally diverse partners. The LIM domain appeared in eukaryotes sometime prior to the most recent common ancestor of plants, fungi, amoeba and animals. In animal cells, LIM domain-containing proteins often shuttle between the cell nucleus where they can regulate gene expression, and the cytoplasm where they are usually associated with actin cytoskeletal structures involved in connecting cells together and to the surrounding matrix, such as stress fibers, focal adhesions and adherens junctions.
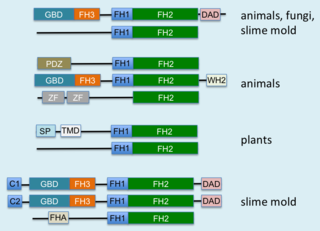
Formins (formin homology proteins) are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end (barbed end) of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

Wiskott–Aldrich syndrome protein family member 2 is a protein that in humans is encoded by the WASF2 gene.
Cytoplasmic protein NCK1 is a protein that in humans is encoded by the NCK1 gene.

Vasodilator-stimulated phosphoprotein is a protein that in humans is encoded by the VASP gene.

Ras GTPase-activating-like protein IQGAP1 (IQGAP1) also known as p195 is a ubiquitously expressed protein that in humans is encoded by the IQGAP1 gene. IQGAP1 is a scaffold protein involved in regulating various cellular processes ranging from organization of the actin cytoskeleton, transcription, and cellular adhesion to regulating the cell cycle.

Protein enabled homolog is a protein that in humans is encoded by the ENAH gene.

CD2-associated protein is a protein that in humans is encoded by the CD2AP gene.

WAS/WASL-interacting protein (WIP) is a protein that in humans is encoded by the WIPF1 gene.

Ena/VASP-like protein is a member of the Ena/VASP family of proteins that in humans is encoded by the EVL gene.

Protein cordon-bleu is a protein that in humans is encoded by the COBL gene.

The WW domain is a modular protein domain that mediates specific interactions with protein ligands. This domain is found in a number of unrelated signaling and structural proteins and may be repeated up to four times in some proteins. Apart from binding preferentially to proteins that are proline-rich, with particular proline-motifs, [AP]-P-P-[AP]-Y, some WW domains bind to phosphoserine- and phosphothreonine-containing motifs.

The Actin assembly-inducing protein (ActA) is a protein encoded and used by Listeria monocytogenes to propel itself through a mammalian host cell. ActA is a bacterial surface protein comprising a membrane-spanning region. In a mammalian cell, the bacterial ActA interacts with the Arp2/3 complex and actin monomers to induce actin polymerization on the bacterial surface generating an actin comet tail. The gene encoding ActA is named actA or prtB.

In molecular biology, the GYF domain is an approximately 60-amino acid protein domain which contains a conserved GP[YF]xxxx[MV]xxWxxx[GN]YF motif. It was identified in the human intracellular protein termed CD2 binding protein 2 (CD2BP2), which binds to a site containing two tandem PPPGHR segments within the cytoplasmic region of CD2. Binding experiments and mutational analyses have demonstrated the critical importance of the GYF tripeptide in ligand binding. A GYF domain is also found in several other eukaryotic proteins of unknown function. It has been proposed that the GYF domain found in these proteins could also be involved in proline-rich sequence recognition. Resolution of the structure of the CD2BP2 GYF domain by NMR spectroscopy revealed a compact domain with a beta-beta-alpha-beta-beta topology, where the single alpha-helix is tilted away from the twisted, anti-parallel beta-sheet. The conserved residues of the GYF domain create a contiguous patch of predominantly hydrophobic nature which forms an integral part of the ligand-binding site. There is limited homology within the C-terminal 20-30 amino acids of various GYF domains, supporting the idea that this part of the domain is structurally but not functionally important.