
Pseudouridine is an isomer of the nucleoside uridine in which the uracil is attached via a carbon-carbon instead of a nitrogen-carbon glycosidic bond.
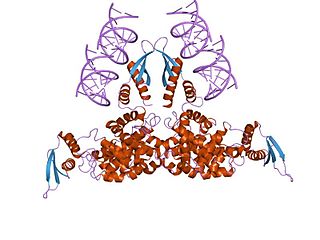
Ribonuclease III (RNase III or RNase C)(BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
In enzymology, a tRNA (uracil-5-)-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA-pseudouridine synthase I is an enzyme that catalyzes the chemical reaction

Helix 69 is a hairpin RNA structure containing 19 nucleotides in large subunit of the ribosome. Ribosome consists of large and small subunits joined with inter subunit bridges. Helix 69 interacts with the helix 44 (h44) of the small subunit to form the largest interface of two subunits called inter-subunit bridge B2a, one of the most conserved regions of the ribosome. Helix 69 is proposed to be a good drug target for antibacterial drugs. Many of the recent crystal structures have shown the involvement of this hairpin in different stages of the protein translation process. By targeting bacterial helix 69 specifically, protein synthesis in bacteria could be halted thus killing the bacteria.
16S rRNA (guanine1207-N2)-methyltransferase (EC 2.1.1.172, m2G1207 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (guanine1207-N2)-methyltransferase. This enzyme catalyses the following chemical reaction
16S rRNA (cytosine967-C5)-methyltransferase (EC 2.1.1.176, rsmB (gene), fmu (gene), 16S rRNA m5C967 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (cytosine967-C5)-methyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (pseudouridine1915-N3)-methyltransferase (EC 2.1.1.177, YbeA, RlmH, pseudouridine methyltransferase, m3Psi methyltransferase, Psi1915-specific methyltransferase, rRNA large subunit methyltransferase H) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (pseudouridine1915-N3)-methyltransferase. This enzyme catalyses the following chemical reaction
16S rRNA pseudouridine516 synthase (EC 5.4.99.19, 16S RNA pseudouridine516 synthase, 16S PsiI516 synthase, 16S RNA Psi516 synthase, RNA pseudouridine synthase RsuA, RsuA, 16S RNA pseudouridine 516 synthase) is an enzyme with systematic name 16S rRNA-uridine516 uracil mutase. This enzyme catalyses the following chemical reaction
23S rRNA pseudouridine2457 synthase is an enzyme with systematic name 23S rRNA-uridine2457 uracil mutase. This enzyme catalyses the following chemical reaction
23S rRNA pseudouridine2604 synthase is an enzyme with systematic name 23S rRNA-uridine2604 uracil mutase. This enzyme catalyses the following chemical reaction
23S rRNA pseudouridine2605 synthase is an enzyme with systematic name 23S rRNA-uridine2605 uracil mutase. This enzyme catalyses the following chemical reaction
23S rRNA pseudouridine1911/1915/1917 synthase (EC 5.4.99.23, RluD, pseudouridine synthase RluD) is an enzyme with systematic name 23S rRNA-uridine1911/1915/1917 uracil mutase. This enzyme catalyses the following chemical reaction
23S rRNA pseudouridine955/2504/2580 synthase is an enzyme with systematic name 23S rRNA-uridine955/2504/2580 uracil mutase. This enzyme catalyses the following chemical reaction
tRNA pseudouridine55 synthase is an enzyme with systematic name tRNA-uridine55 uracil mutase. This enzyme catalyses the following chemical reaction
tRNA pseudouridine65 synthase is an enzyme with systematic name tRNA-uridine65 uracil mutase. This enzyme catalyses the following chemical reaction
tRNA pseudouridine13 synthase is an enzyme with systematic name tRNA-uridine13 uracil mutase. This enzyme catalyses the following chemical reaction
tRNA pseudouridine32 synthase is an enzyme with systematic name tRNA-uridine32 uracil mutase. This enzyme catalyses the following chemical reaction
tRNA pseudouridine38/39 synthase is an enzyme with systematic name tRNA-uridine38/39 uracil mutase. This enzyme catalyses the following chemical reaction

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.




