
Caenorhabditis elegans is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like) and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans. Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.

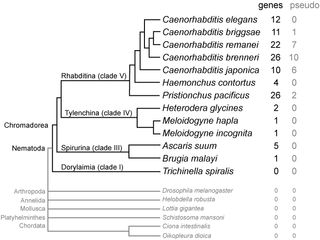
Comparative genomics is a branch of biological research that examines genome sequences across a spectrum of species, spanning from humans and mice to a diverse array of organisms from bacteria to chimpanzees. This large-scale holistic approach compares two or more genomes to discover the similarities and differences between the genomes and to study the biology of the individual genomes. Comparison of whole genome sequences provides a highly detailed view of how organisms are related to each other at the gene level. By comparing whole genome sequences, researchers gain insights into genetic relationships between organisms and study evolutionary changes. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, Comparative genomics provides a powerful tool for studying evolutionary changes among organisms, helping to identify genes that are conserved or common among species, as well as genes that give unique characteristics of each organism. Moreover, these studies can be performed at different levels of the genomes to obtain multiple perspectives about the organisms.

Mitochondrial 5-demethoxyubiquinone hydroxylase, also known as coenzyme Q7, hydroxylase, is an enzyme that in humans is encoded by the COQ7 gene. The clk-1 (clock-1) gene encodes this protein that is necessary for ubiquinone biosynthesis in the worm Caenorhabditis elegans and other eukaryotes. The mouse version of the gene is called mclk-1 and the human, fruit fly and yeast homolog COQ7.
WormBook is an open access, comprehensive collection of original, peer-reviewed chapters covering topics related to the biology of the nematode worm Caenorhabditis elegans . WormBook also includes WormMethods, an up-to-date collection of methods and protocols for C. elegans researchers.

Caenorhabditis is a genus of nematodes which live in bacteria-rich environments like compost piles, decaying dead animals and rotting fruit. The name comes from Greek: caeno- ; rhabditis = rod-like.

Most animal testing involves invertebrates, especially Drosophila melanogaster, a fruit fly, and Caenorhabditis elegans, a nematode. These animals offer scientists many advantages over vertebrates, including their short life cycle, simple anatomy and the ease with which large numbers of individuals may be studied. Invertebrates are often cost-effective, as thousands of flies or nematodes can be housed in a single room.
SmY ribonucleic acids are a family of small nuclear RNAs found in some species of nematode worms. They are thought to be involved in mRNA trans-splicing.
The nematode worm Caenorhabditis elegans was first studied in the laboratory by Victor Nigon and Ellsworth Dougherty in the 1940s, but came to prominence after being adopted by Sydney Brenner in 1963 as a model organism for the study of developmental biology using genetics. In 1974, Brenner published the results of his first genetic screen, which isolated hundreds of mutants with morphological and functional phenotypes, such as being uncoordinated. In the 1980s, John Sulston and co-workers identified the lineage of all 959 cells in the adult hermaphrodite, the first genes were cloned, and the physical map began to be constructed. In 1998, the worm became the first multi-cellular organism to have its genome sequenced. Notable research using C. elegans includes the discoveries of caspases, RNA interference, and microRNAs. Eight scientists have won the Nobel Prize for their work on C. elegans.
The Rhabditidae are a family of nematodes which includes the model organism Caenorhabditis elegans.
DAF-16 is the sole ortholog of the FOXO family of transcription factors in the nematode Caenorhabditis elegans. It is responsible for activating genes involved in longevity, lipogenesis, heat shock survival and oxidative stress responses. It also protects C.elegans during food deprivation, causing it to transform into a hibernation - like state, known as a Dauer. DAF-16 is notable for being the primary transcription factor required for the profound lifespan extension observed upon mutation of the insulin-like receptor DAF-2. The gene has played a large role in research into longevity and the insulin signalling pathway as it is located in C. elegans, a successful ageing model organism.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.

Caenorhabditis elegans- microbe interactions are defined as any interaction that encompasses the association with microbes that temporarily or permanently live in or on the nematode C. elegans. The microbes can engage in a commensal, mutualistic or pathogenic interaction with the host. These include bacterial, viral, unicellular eukaryotic, and fungal interactions. In nature C. elegans harbours a diverse set of microbes. In contrast, C. elegans strains that are cultivated in laboratories for research purposes have lost the natural associated microbial communities and are commonly maintained on a single bacterial strain, Escherichia coli OP50. However, E. coli OP50 does not allow for reverse genetic screens because RNAi libraries have only been generated in strain HT115. This limits the ability to study bacterial effects on host phenotypes. The host microbe interactions of C. elegans are closely studied because of their orthologs in humans. Therefore, the better we understand the host interactions of C. elegans the better we can understand the host interactions within the human body.
Caenorhabditis tropicalis is a species of Caenorhabditis nematodes, belonging to the Elegans super-group and Elegans group within the genus. It is a close relative of C. wallacei.C. tropicalis is collected frequently in tropical South America, Caribbean islands, and various islands in the Indian and Pacific Oceans from rotting fruit, flowers and stems. C. tropicalis was referred to as "C. sp. 11" prior to 2014.
Caenorhabditis nigoni is a male-female species in the Elegans group of the genus Caenorhabditis, first identified and described as "Caenorhabditis species 9" or "C. sp. 9" before being renamed as "C. nigoni". The specific epithet is a tribute to Victor Nigon who first studied Caenorhabditis elegans in the laboratory with Ellsworth Dougherty in the 1940s. Isolates come from the Democratic Republic of the Congo and Kerala, India.
Victor Marc Nigon was a biologist who was first to study the nematode worm Caenorhabditis elegans in the laboratory, with Ellsworth Dougherty, in the 1940s.
Ellsworth C. Dougherty was a biologist who was first to study the nematode worm Caenorhabditis elegans in the laboratory, with Victor Nigon, in the 1940s. He did most of his studies and medical work in California.

Caenorhabditis sinica, is a species of Caenorhabditis nematodes, belonging to the Elegans super-group and Elegans group within the genus. It is closely related to several species isolated from the lands adjacent to the Indian and Pacific Oceans, as well as to C. briggsae and C. nigoni. The species was known as “C. sp. 5” prior to 2014. C. sinica is known for having very high genetic diversity in its genome. Like other Caenorhabditis species, C. sinica is a ~1mm long roundworm with a transparent cuticle and that eats bacteria. Wild isolate strains of C. sinica have been collected from various rotting plant tissue substrates in temperate and tropical regions throughout China since its initial isolation in 2005.
The G-value paradox arises from the lack of correlation between the number of protein-coding genes among eukaryotes and their relative biological complexity. The microscopic nematode Caenorhabditis elegans, for example, is composed of only a thousand cells but has about the same number of genes as a human. Researchers suggest resolution of the paradox may lie in mechanisms such as alternative splicing and complex gene regulation that make the genes of humans and other complex eukaryotes relatively more productive.
Paul W. Sternberg is an American biologist. He does research for WormBase on C. elegans, a model organism.

Warwick Llewellyn Nicholas (1926–2010) was an Australian zoologist known as a pioneer in the field of nematology. He was a foundational member of the Australian Society for Parasitology (ASP) and in 1964, he organised the first ASP meeting. He became President of the Society in 1978, before being an elected Fellow from 1979.








