
Sulfur (also spelled sulphur in British English) is a chemical element; it has symbol S and atomic number 16. It is abundant, multivalent and nonmetallic. Under normal conditions, sulfur atoms form cyclic octatomic molecules with the chemical formula S8. Elemental sulfur is a bright yellow, crystalline solid at room temperature.

Methionine is an essential amino acid in humans.
Mycoplasma pneumoniae is a species of very small cell bacteria that lack a cell wall, in the class Mollicutes. M. pneumoniae is a human pathogen that causes the disease Mycoplasma pneumonia, a form of atypical bacterial pneumonia related to cold agglutinin disease. M. pneumoniae is characterized by the absence of a peptidoglycan cell wall and resulting resistance to many cell wall active antibacterial agents.
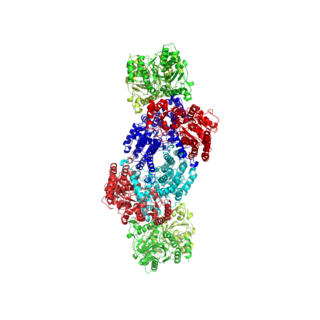
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.
Iron(II) sulfide or ferrous sulfide is one of a family of chemical compounds and minerals with the approximate formula FeS. Iron sulfides are often iron-deficient non-stoichiometric. All are black, water-insoluble solids.

Sulfur assimilation is the process by which living organisms incorporate sulfur into their biological molecules. In plants, sulfate is absorbed by the roots and then transported to the chloroplasts by the transipration stream where the sulfur are reduced to sulfide with the help of a series of enzymatic reactions. Furthermore, the reduced sulfur is incorporated into cysteine, an amino acid that is a precursor to many other sulfur-containing compounds. In animals, sulfur assimilation occurs primarily through the diet, as animals cannot produce sulfur-containing compounds directly. Sulfur is incorporated into amino acids such as cysteine and methionine, which are used to build proteins and other important molecules.
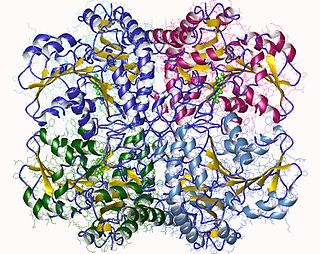
The enzyme cystathionine γ-lyase (EC 4.4.1.1, CTH or CSE; also cystathionase; systematic name L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming)) breaks down cystathionine into cysteine, 2-oxobutanoate (α-ketobutyrate), and ammonia:
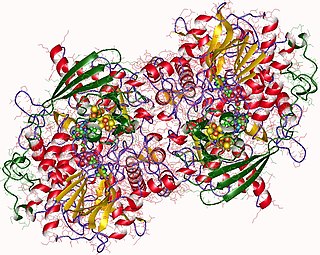
Adenylyl-sulfate reductase is an enzyme that catalyzes the chemical reaction of the reduction of adenylyl-sulfate/adenosine-5'-phosphosulfate (APS) to sulfite through the use of an electron donor cofactor. The products of the reaction are AMP and sulfite, as well as an oxidized electron donor cofactor.
The enzyme L-3-cyanoalanine synthase catalyzes the chemical reaction

In enzymology, a 3-mercaptopyruvate sulfurtransferase is an enzyme that catalyzes the chemical reactions of 3-mercaptopyruvate. This enzyme belongs to the family of transferases, specifically the sulfurtransferases. This enzyme participates in cysteine metabolism. It is encoded by the MPST gene.
Biotin synthase (BioB) is an enzyme that catalyzes the conversion of dethiobiotin (DTB) to biotin; this is the final step in the biotin biosynthetic pathway. Biotin, also known as vitamin B7, is a cofactor used in carboxylation, decarboxylation, and transcarboxylation reactions in many organisms including humans. Biotin synthase is an S-Adenosylmethionine (SAM) dependent enzyme that employs a radical mechanism to thiolate dethiobiotin, thus converting it to biotin.
In enzymology, a tRNA sulfurtransferase is an enzyme that catalyzes the chemical reaction

Cysteine desulfurase, mitochondrial is an enzyme that in humans is encoded by the NFS1 gene.
Sulfur is metabolized by all organisms, from bacteria and archaea to plants and animals. Sulfur can have an oxidation state from -2 to +6 and is reduced or oxidized by a diverse range of organisms. The element is present in proteins, sulfate esters of polysaccharides, steroids, phenols, and sulfur-containing coenzymes.

In biochemistry, the iron–sulfur cluster biosynthesis describes the components and processes involved in the biosynthesis of iron–sulfur proteins. The topic is of interest because these proteins are pervasive. The iron sulfur proteins contain iron–sulfur clusters, some with elaborate structures, that feature iron and sulfide centers. One broad biosynthetic task is producing sulfide (S2-), which requires various families of enzymes. Another broad task is affixing the sulfide to iron, which is achieved on scaffolds, which are nonfunctional. Finally these Fe-S cluster is transferred to a target protein, which then become functional.
Molybdenum cofactor sulfurtransferase (EC 2.8.1.9, molybdenum cofactor sulfurase, ABA3, MoCo sulfurase, MoCo sulfurtransferase) is an enzyme with systematic name L-cysteine:molybdenum cofactor sulfurtransferase. This enzyme catalyses the following chemical reaction
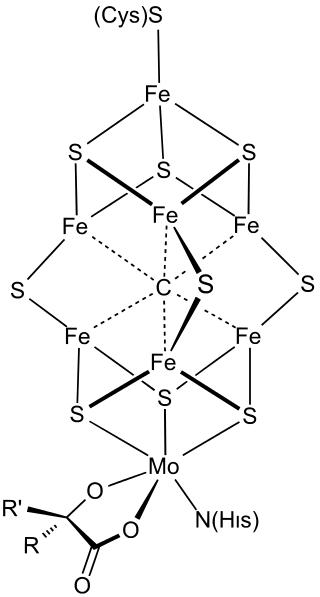
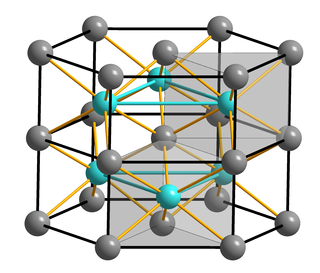
FeMoco (FeMo cofactor) is the primary cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Because it contains iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C.
Dissimilatory sulfite reductase is an enzyme that participates in sulfur metabolism in dissimilatory sulfate reduction.

Isethionate sulfite-lyase is a glycyl radical enzyme that catalyzes the degradation of isethionate into acetaldehyde and sulfite through the cleavage of a carbon-sulfur bond. This conversion is a necessary step for taurine catabolism in anaerobic bacteria like Bilophila wadsworthia. IslA is activated by the enzyme IslB which uses S-adenoslymethionine (SAM) as the initial radical donor.













