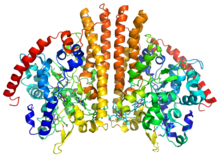
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.
An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. Many of the enzymes in the electron transport chain are embedded within the membrane.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
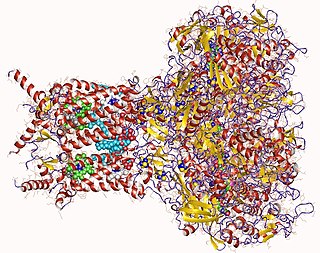
Formate dehydrogenases are a set of enzymes that catalyse the oxidation of formate to carbon dioxide, donating the electrons to a second substrate, such as NAD+ in formate:NAD+ oxidoreductase (EC 1.17.1.9) or to a cytochrome in formate:ferricytochrome-b1 oxidoreductase (EC 1.2.2.1). This family of enzymes has attracted attention as inspiration or guidance on methods for the carbon dioxide fixation, relevant to global warming.
Nitrite oxidoreductase is an enzyme involved in nitrification. It is the last step in the process of aerobic ammonia oxidation, which is carried out by two groups of nitrifying bacteria: ammonia oxidizers such as Nitrosospira, Nitrosomonas, and Nitrosococcus convert ammonia to nitrite, while nitrite oxidizers such as Nitrobacter and Nitrospira oxidize nitrite to nitrate. NXR is responsible for producing almost all nitrate found in nature.

Nitric oxide dioxygenase (EC 1.14.12.17) is an enzyme that catalyzes the conversion of nitric oxide (NO) to nitrate (NO−
3) . The net reaction for the reaction catalyzed by nitric oxide dioxygenase is shown below:
In enzymology, a quinoprotein glucose dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, an ethylbenzene hydroxylase (EC 1.17.99.2) is an enzyme that catalyzes the chemical reaction
In enzymology, an iron—cytochrome-c reductase (created 1972 as EC 1.9.99.1, transferred 2014 to EC 1.9.98.1) is an enzyme that catalyzes the chemical reaction

In enzymology, a NADPH—hemoprotein reductase is an enzyme that catalyzes the chemical reaction
Nitric oxide reductase, an enzyme, catalyzes the reduction of nitric oxide (NO) to nitrous oxide (N2O). The enzyme participates in nitrogen metabolism and in the microbial defense against nitric oxide toxicity. The catalyzed reaction may be dependent on different participating small molecules: Cytochrome c (EC: 1.7.2.5, Nitric oxide reductase (cytochrome c)), NADPH (EC:1.7.1.14), or Menaquinone (EC:1.7.5.2).
In enzymology, a nitrite reductase [NAD(P)H] (EC 1.7.1.4) is an enzyme that catalyzes the chemical reaction
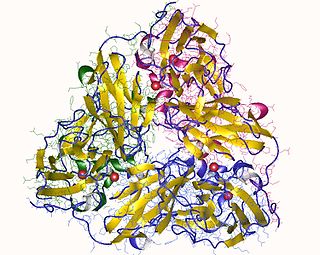
In enzymology, a nitrite reductase (NO-forming) (EC 1.7.2.1) is an enzyme that catalyzes the chemical reaction

In enzymology, a nitrous oxide reductase also known as nitrogen:acceptor oxidoreductase (N2O-forming) is an enzyme that catalyzes the final step in bacterial denitrification, the reduction of nitrous oxide to dinitrogen.

Thiosulfate dehydrogenase is an enzyme that catalyzes the chemical reaction:

Flavocytochrome c sulfide dehydrogenase, also known as Sulfide-cytochrome-c reductase (flavocytochrome c) (EC 1.8.2.3), is an enzyme with systematic name hydrogen-sulfide:flavocytochrome c oxidoreductase. It is found in sulfur-oxidising bacteria such as the purple phototrophic bacteria Allochromatium vinosum. This enzyme catalyses the following chemical reaction:

Fumarate reductase (quinol) (EC 1.3.5.4, QFR,FRD, menaquinol-fumarate oxidoreductase, quinol:fumarate reductase) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:
Pyruvate dehydrogenase (quinone) (EC 1.2.5.1, pyruvate dehydrogenase, pyruvic dehydrogenase, pyruvic (cytochrome b1) dehydrogenase, pyruvate:ubiquinone-8-oxidoreductase, pyruvate oxidase (ambiguous)) is an enzyme with systematic name pyruvate:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction
Nitrate reductase (quinone) (EC 1.7.5.1, nitrate reductase A, nitrate reductase Z, quinol/nitrate oxidoreductase, quinol-nitrate oxidoreductase, quinol:nitrate oxidoreductase, NarA, NarZ, NarGHI) is an enzyme with systematic name nitrite:quinone oxidoreductase. This enzyme catalyses the following chemical reaction

Cytochrome d, previously known as cytochrome a2, is a name for all cytochromes that contain heme D as a cofactor. Two unrelated classes of cytochrome d are known: Cytochrome bd, an enzyme that generates a charge across the membrane so that protons will move, and cytochrome cd1, a nitrite reductase.