UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, USA.
The European Bioinformatics Institute (EMBL-EBI) is an intergovernmental organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wellcome Genome Campus in Hinxton near Cambridge, and employs over 600 full-time equivalent (FTE) staff.
The Bioinformatic Harvester was a bioinformatic meta search engine created by the European Molecular Biology Laboratory and subsequently hosted and further developed by KIT Karlsruhe Institute of Technology for genes and protein-associated information. Harvester currently works for human, mouse, rat, zebrafish, drosophila and arabidopsis thaliana based information. Harvester cross-links >50 popular bioinformatic resources and allows cross searches. Harvester serves tens of thousands of pages every day to scientists and physicians. Since 2014 the service is down.

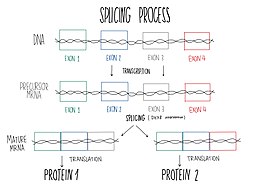
The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box (SF2) helicases, which are involved in RNA metabolism.

Spliceosome RNA helicase BAT1 is an enzyme that in humans is encoded by the BAT1 gene.

Probable ATP-dependent RNA helicase DDX17 (p72) is an enzyme that in humans is encoded by the DDX17 gene.

Splicing factor 3A subunit 2 is a protein that in humans is encoded by the SF3A2 gene.
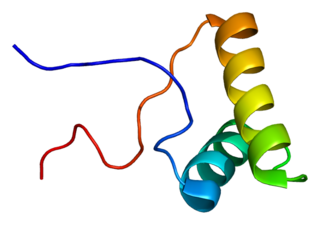
Splicing factor 3B subunit 2 is a protein that in humans is encoded by the SF3B2 gene.
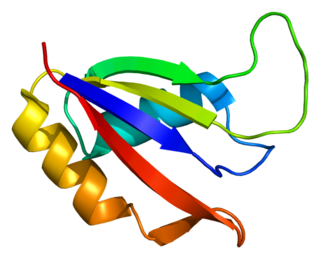
Aly/REF export factor, also known as THO complex subunit 4 is a protein that in humans is encoded by the ALYREF gene.

Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 is an enzyme that in humans is encoded by the DHX38 gene.

Chromodomain-helicase-DNA-binding protein 8 is an enzyme that in humans is encoded by the CHD8 gene.

ATP-dependent RNA helicase DDX19B is an enzyme that in humans is encoded by the DDX19B gene.

Probable ATP-dependent RNA helicase DDX23 is an enzyme that in humans is encoded by the DDX23 gene.

Probable ATP-dependent RNA helicase DHX36 also known as DEAH box protein 36 (DHX36) or MLE-like protein 1 (MLEL1) or G4 resolvase 1 (G4R1) or RNA helicase associated with AU-rich elements (RHAU) is an enzyme that in humans is encoded by the DHX36 gene.

Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 is an enzyme that in humans is encoded by the DHX15 gene.

Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX32 is an enzyme that in humans is encoded by the DHX32 gene.

Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 is an enzyme that in humans is encoded by the DHX16 gene.

PIF1 5'-to-3' DNA helicase is a protein that in humans is encoded by the PIF1 gene.

MIF4GD, or MIF4G domain-containing protein, is a protein which in humans is encoded by the MIF4GD gene. It is also known as SLIP1, SLBP -interacting protein 1, AD023, and MIFD. MIF4GD is expressed ubiquitously in humans, and has been found to be involved in activating proteins for histone mRNA translation, alternative splicing and translation of mRNAs, and is a factor in the regulation of cell proliferation.