Related Research Articles

Escherichia coli ( ESH-ə-RIK-ee-ə KOH-ly) is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.

DNA polymerase II is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
Christopher Francis Higgins is a British molecular biologist, geneticist, academic and scientific advisor. He was the Vice-Chancellor of Durham University from 2007 to 2014. He took early retirement on 30 September 2014, following a discussion at Senate on limiting the powers of the Vice Chancellor. He was previously the director of the MRC Clinical Sciences Centre and Head of Division in the Faculty of Medicine at Imperial College London.
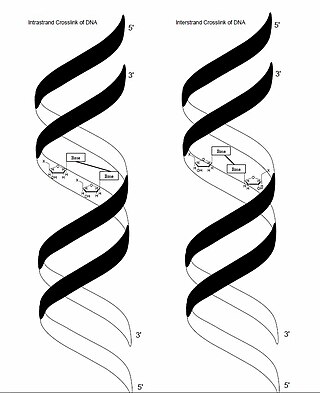
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.
AlkB (Alkylation B) is a protein found in E. coli, induced during an adaptive response and involved in the direct reversal of alkylation damage. AlkB specifically removes alkylation damage to single stranded (SS) DNA caused by SN2 type of chemical agents. It efficiently removes methyl groups from 1-methyl adenines, 3-methyl cytosines in SS DNA. AlkB is an alpha-ketoglutarate-dependent hydroxylase, a superfamily non-haem iron-containing proteins. It oxidatively demethylates the DNA substrate. Demethylation by AlkB is accompanied with release of CO2, succinate, and formaldehyde.

DNA-3-methyladenine glycosylase also known as 3-alkyladenine DNA glycosylase (AAG) or N-methylpurine DNA glycosylase (MPG) is an enzyme that in humans is encoded by the MPG gene.

Endonuclease III-like protein 1 is an enzyme that in humans is encoded by the NTHL1 gene.
Endonuclease VIII-like 1 is an enzyme that in humans is encoded by the NEIL1 gene.
In DNA repair, the Ada regulon is a set of genes whose expression is essential to adaptive response, which is triggered in prokaryotic cells by exposure to sub-lethal doses of alkylating agents. This allows the cells to tolerate the effects of such agents, which are otherwise toxic and mutagenic.
DNA oxidative demethylase (EC 1.14.11.33, alkylated DNA repair protein, alpha-ketoglutarate-dependent dioxygenase ABH1, alkB (gene)) is an enzyme with systematic name methyl DNA-base, 2-oxoglutarate:oxygen oxidoreductase (formaldehyde-forming). This enzyme catalyses the following chemical reaction
Deoxyribodipyrimidine endonucleosidase is an enzyme with systematic name deoxy-D-ribocyclobutadipyrimidine polynucleotidodeoxyribohydrolase. This enzyme catalyses the following chemical reaction
DNA-3-methyladenine glycosylase II is an enzyme that catalyses the following chemical reaction:
DNA-formamidopyrimidine glycosylase is an enzyme with systematic name DNA glycohydrolase . FPG is a base excision repair enzyme which recognizes and removes a wide range of oxidized purines from correspondingly damaged DNA. It was discovered by Zimbabwean scientist Christopher J. Chetsanga in 1975.
Double-stranded uracil-DNA glycosylase is an enzyme with systematic name uracil-double-stranded DNA deoxyribohydrolase (uracil-releasing). This enzyme catalyses a specific chemical reaction: it hydrolyses mismatched double-stranded DNA and polynucleotides, releasing free uracil.

AlkD is an enzyme belonging to a family of DNA glycosylases that are involved in DNA repair. It was discovered by a team of Norwegian biologists from Oslo in 2006. It was isolated from a soil-dwelling Gram-positive bacteria Bacillus cereus, along with another enzyme AlkC. AlkC and AlkD are most probably derived from the same protein as indicated by their close resemblance. They are also found in other prokaryotes. Among eukaryotes, they are found only in the single-celled species only, such as Entamoeba histolytica and Dictyostelium discoideum. The enzyme specifically targets 7mG (methyl-guanine) in the DNA, and is, therefore, unique among DNA glycosylases. It can also act on other methylpurines with less affinity. It indicates that the enzyme is specific for locating and cutting (excision) of chemically modified bases from DNA, exactly at 7mG, whenever there are errors in replication. It accelerates the rate of 7mG hydrolysis 100-fold over the spontaneous depurination. Thus, it protects the genome from harmful changes induced by chemical and environmental agents. Its crystal structure was described in 2008. It is the first HEAT repeat protein identified to interact with nucleic acids or to contain enzymatic activity.

Christopher J. Chetsanga is a prominent Zimbabwean scientist who is a member of the African Academy of Sciences and The World Academy of Sciences. He discovered two enzymes involved in DNA repair. He has also held various academic administrative posts like Vice-Chancellor, Director and Dean.
References
- ↑ Evensen G, Seeberg E (April 1982). "Adaptation to alkylation resistance involves the induction of a DNA glycosylase". Nature. 296 (5859): 773–5. Bibcode:1982Natur.296..773E. doi:10.1038/296773a0. PMID 7040984. S2CID 4318955.
- ↑ Karran P, Hjelmgren T, Lindahl T (April 1982). "Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents". Nature. 296 (5859): 770–3. Bibcode:1982Natur.296..770K. doi:10.1038/296770a0. PMID 7040983. S2CID 4367726.
- ↑ Thomas L, Yang CH, Goldthwait DA (March 1982). "Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine". Biochemistry. 21 (6): 1162–9. doi:10.1021/bi00535a009. PMID 7041972.