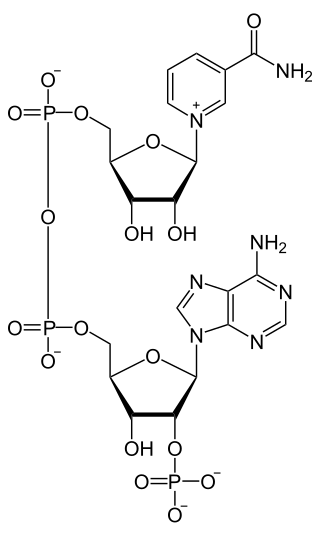
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form, whereas NADP+ is the oxidized form. NADP+ is used by all forms of cellular life.
Acyl-CoA dehydrogenases (ACADs) are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β-oxidation in the mitochondria of cells. Their action results in the introduction of a trans double-bond between C2 (α) and C3 (β) of the acyl-CoA thioester substrate. Flavin adenine dinucleotide (FAD) is a required co-factor in addition to the presence of an active site glutamate in order for the enzyme to function.

Cytochrome P450 reductase is a membrane-bound enzyme required for electron transfer from NADPH to cytochrome P450 and other heme proteins including heme oxygenase in the endoplasmic reticulum of the eukaryotic cell.

Electron-transferring-flavoprotein dehydrogenase is an enzyme that transfers electrons from electron-transferring flavoprotein in the mitochondrial matrix, to the ubiquinone pool in the inner mitochondrial membrane. It is part of the electron transport chain. The enzyme is found in both prokaryotes and eukaryotes and contains a flavin and FE-S cluster. In humans, it is encoded by the ETFDH gene. Deficiency in ETF dehydrogenase causes the human genetic disease multiple acyl-CoA dehydrogenase deficiency.
In enzymology, sarcosine dehydrogenase (EC 1.5.8.3) is a mitochondrial enzyme that catalyzes the chemical reaction N-demethylation of sarcosine to give glycine. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donor with other acceptors. The systematic name of this enzyme class is sarcosine:acceptor oxidoreductase (demethylating). Other names in common use include sarcosine N-demethylase, monomethylglycine dehydrogenase, and sarcosine:(acceptor) oxidoreductase (demethylating). Sarcosine dehydrogenase is closely related to dimethylglycine dehydrogenase, which catalyzes the demethylation reaction of dimethylglycine to sarcosine. Both sarcosine dehydrogenase and dimethylglycine dehydrogenase use FAD as a cofactor. Sarcosine dehydrogenase is linked by electron-transferring flavoprotein (ETF) to the respiratory redox chain. The general chemical reaction catalyzed by sarcosine dehydrogenase is:
In enzymology, a cholest-5-ene-3β,7α-diol 3β-dehydrogenase (EC 1.1.1.181) is an enzyme that catalyzes the chemical reaction

In enzymology, an alcohol oxidase (EC 1.1.3.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a cellobiose dehydrogenase (acceptor) (EC 1.1.99.18) is an enzyme that catalyzes the chemical reaction

In enzymology, a cholesterol oxidase (EC 1.1.3.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a choline dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, a choline oxidase (EC 1.1.3.17) is an enzyme that catalyzes the chemical reaction
In enzymology, a quinoprotein glucose dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, a thiamine oxidase (EC 1.1.3.23) is an enzyme that catalyzes the chemical reaction

In enzymology, an L-amino acid oxidase (LAAO) (EC 1.4.3.2) is an enzyme that catalyzes the chemical reaction.
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.
The aldehyde oxidase and xanthine dehydrogenase, a/b hammerhead domain is an evolutionary conserved protein domain.
The carboxypeptidase A family can be divided into two subfamilies: carboxypeptidase H (regulatory) and carboxypeptidase A (digestive). Members of the H family have longer C-termini than those of family A, and carboxypeptidase M is bound to the membrane by a glycosylphosphatidylinositol anchor, unlike the majority of the M14 family, which are soluble.

In molecular biology, the FAD dependent oxidoreductase family of proteins is a family of FAD dependent oxidoreductases. Members of this family include Glycerol-3-phosphate dehydrogenase EC 1.1.99.5, Sarcosine oxidase beta subunit EC 1.5.3.1, D-amino-acid dehydrogenase EC 1.4.99.1, D-aspartate oxidase EC 1.4.3.1.

Fumarate reductase (quinol) (EC 1.3.5.4, QFR,FRD, menaquinol-fumarate oxidoreductase, quinol:fumarate reductase) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:
Alice Vrielink is a structural biologist and Professor of Structural Biology in the School of Molecular Sciences at the University of Western Australia. She is known for her work determining the structures of macromolecules such as enzymes and nucleic acids.











