Related Research Articles
Haplogroup K, formerly Haplogroup UK, is a human mitochondrial DNA (mtDNA) haplogroup. It is defined by the HVR1 mutations 16224C and 16311C. It is now known that K is a subclade of U8.
Haplogroup J is a human mitochondrial DNA (mtDNA) haplogroup. The clade derives from the haplogroup JT, which also gave rise to haplogroup T. Within the field of medical genetics, certain polymorphisms specific to haplogroup J have been associated with Leber's hereditary optic neuropathy.
Haplogroup L2 is a human mitochondrial DNA (mtDNA) haplogroup with a widespread modern distribution, particularly in Subequatorial Africa. Its L2a subclade is a somewhat frequent and widely distributed mtDNA cluster on the continent, as well as among those in the Americas.

Haplogroup L1 is a human mitochondrial DNA (mtDNA) haplogroup. It is most common in Central Africa and West Africa. It diverged from L1-6 at about 140,000 years ago . Its emergence is associated with the early peopling of Africa by anatomically modern humans during the Eemian, and it is now mostly found in African pygmies.
Haplogroup I is a human mitochondrial DNA (mtDNA) haplogroup. It is believed to have originated about 21,000 years ago, during the Last Glacial Maximum (LGM) period in West Asia. The haplogroup is unusual in that it is now widely distributed geographically, but is common in only a few small areas of East Africa, West Asia and Europe. It is especially common among the El Molo and Rendille peoples of Kenya, various regions of Iran, the Lemko people of Slovakia, Poland and Ukraine, the island of Krk in Croatia, the department of Finistère in France and some parts of Scotland and Ireland.

In human genetics, a human mitochondrial DNA haplogroup is a haplogroup defined by differences in human mitochondrial DNA. Haplogroups are used to represent the major branch points on the mitochondrial phylogenetic tree. Understanding the evolutionary path of the female lineage has helped population geneticists trace the matrilineal inheritance of modern humans back to human origins in Africa and the subsequent spread around the globe.
Haplogroup JT is a human mitochondrial DNA (mtDNA) haplogroup.
In human mitochondrial genetics, haplogroup E is a human mitochondrial DNA (mtDNA) haplogroup typical for the Malay Archipelago. It is a subgroup of haplogroup M9.
Haplogroup L0 is a human mitochondrial DNA (mtDNA) haplogroup.

In human mitochondrial genetics, haplogroup Q is a human mitochondrial DNA (mtDNA) haplogroup typical for Oceania. It is a subgroup of haplogroup M29'Q.
In human mitochondrial genetics, the Haplogroup CZ is a human mitochondrial DNA (mtDNA) haplogroup.
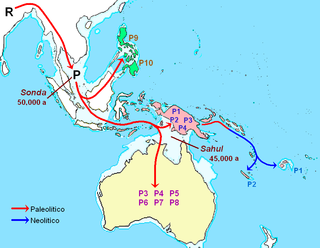
In human mitochondrial genetics, Haplogroup P is a human mitochondrial DNA (mtDNA) haplogroup.

In human genetics, Haplogroup S is a human mitochondrial DNA (mtDNA) haplogroup found only among Indigenous Australians. It is a descendant of macrohaplogroup N.

Haplogroup L4 is a human mitochondrial DNA (mtDNA) haplogroup. It is a small maternal clade primarily restricted to Africa.
In human mitochondrial genetics, Haplogroup L6 is a human mitochondrial DNA (mtDNA) haplogroup. It is a small African haplogroup.

Haplogroup N1a is a human mitochondrial DNA (mtDNA) haplogroup.
In human mitochondrial genetics, Haplogroup H5 is a human mitochondrial DNA (mtDNA) haplogroup descended from Haplogroup H (mtDNA). H5 is defined by T16304C in the HVR1 region and 456 in the HVR2 region.
Haplogroup R0 is a human mitochondrial DNA (mtDNA) haplogroup.
In human mitochondrial genetics, Haplogroup K1a1b1a is a human mitochondrial DNA (mtDNA) haplogroup.
In human mitochondrial genetics, Haplogroup M8 is a human mitochondrial DNA (mtDNA) haplogroup.
References
- ↑ Soares, P; Ermini, L; Thomson, N; Mormina, M; Rito, T; Röhl, A; Salas, A; Oppenheimer, S; MacAulay, V (2009). "Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock". American Journal of Human Genetics. 84 (6): 740–59. doi:10.1016/j.ajhg.2009.05.001. PMC 2694979 . PMID 19500773.
- 1 2 van Oven, Mannis; Manfred Kayser (13 Oct 2008). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–E394. doi: 10.1002/humu.20921 . PMID 18853457. S2CID 27566749.
- ↑ Sarah A. Tishkoff et al. 2007, History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation. Molecular Biology and Evolution 2007 24(10):2180-2195
- ↑ Stevanovitch, A; A. Gilles; E. Bouzaid; R. Kefi; F. Paris; R. P. Gayraud; J. L. Spadoni; F. El-Chenawi; E. Béraud-Colomb (29 Jan 2004). "Mitochondrial DNA Sequence Diversity in a Sedentary Population from Egypt". Annals of Human Genetics. 68 (1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828. S2CID 44901197.
- ↑ Abu-Amero et al. 2008 February. "Mitochondrial DNA structure in the Arabian Peninsula", BMC Evolutionary Biology. 8(45): 52.
- ↑ Sirak, Kendra; Frenandes, Daniel; Novak, Mario; Van Gerven, Dennis; Pinhasi, Ron (2016). "Abstract Book of the IUAES Inter-Congress 2016 - A community divided? Revealing the community genome(s) of Medieval Kulubnarti using next- generation sequencing". Abstract Book of the Iuaes Inter-Congress 2016. IUAES: 115.